-
Notifications
You must be signed in to change notification settings - Fork 0
Computing
To get access to the computing cluster, send Nathan Skene an email with your username and he will add you.
The Imperial CX1 cluster uses PBS as a job manager. PBS has many versions and it cannot neccesarily be assumed that a function you find in an online manual will work exactly as described there. The functions which work on the cluster are best found by typing man qstat while logged to an interactive session.
There is a weekly HPC clinic. I strongly recommend making use of this. You can turn up and experts will help you. Even if you are just unsure about something, go and speak to them. They are held in South Kensington but it's well worth going.
Imperial regularly runs a beginner's guide to high performance computing course. If you have not previously used HPC you'll want to register for this as soon as possible. This can be done through their website:
Imperial also runs a course on software carpentry. If you are unfamiliar with usage of Git and Linux then you should take this course.
Combiz Khozoie wrote useful notes on using the HPC (how to login etc):
The job sizing requirements for Imperial's HPC have changed over time, so please see here for the most updated version: https://wiki.imperial.ac.uk/pages/viewpage.action?spaceKey=HPC&title=New+Job+sizing+guidance
You can request/launch Rstudio containers with a certain amount of resources using HPC's On-Demand service here: https://openondemand.rcs.ic.ac.uk/
For further documentation, please see here: https://wiki.imperial.ac.uk/display/HPC/Open+OnDemand
VPN into the network, then connect with
ssh <YOUR_USERNAME>@login.hpc.ic.ac.uk
To setup ssh on your computer for accessing the cluster add the following to ~/.ssh/config.
Make sure to replace all places marker <YOUR_USERNAME> with your actual Imperial HPC username (without the "< >"):
Host *
AddKeysToAgent yes
IdentityFile ~/.ssh/id_rsa
Host imperial
User <YOUR_USERNAME>
AddKeysToAgent yes
HostName login.cx1.hpc.imperial.ac.uk
ForwardX11Trusted yes
ForwardX11 yes
HostKeyAlgorithms=+ssh-dss
Host imperial-7
User <YOUR_USERNAME>
AddKeysToAgent yes
HostName login-7.cx1.hpc.imperial.ac.uk
ForwardX11Trusted yes
ForwardX11 yes
HostKeyAlgorithms=+ssh-dss
If you use imperial-7 to login then you'll always connect to the same login node which makes using screen/tmux easier.
Rather than entering your password every time you ssh login, you can simply use ssh-keygen to do this once:
ssh-copy-id <YOUR_USERNAME>@login.hpc.ic.ac.uk
This will ask you for your HPC password. Afterwards you will no longer have to enter your password from your local computer. For a more detailed explanation, see here.
You can mount HPC on your local computer, allowing you to easily browse, edit, and import anything that is stored on HPC. You can even open R projects and use your local Rstudio to edit and run code.
Note that this will be slower than running the same code directly on the HPC, as all the data has to be first transferred from HPC to your local computer every time you run a command. Also, all processes will be executed on your machine, meaning you do not have direct access to HPC's compute power through this approach (unless you're submitting pbs job scripts).
Set this up using the following steps:
- On your Desktop, use the hotkey command
CMD+Kto open the following Connect to Server window.
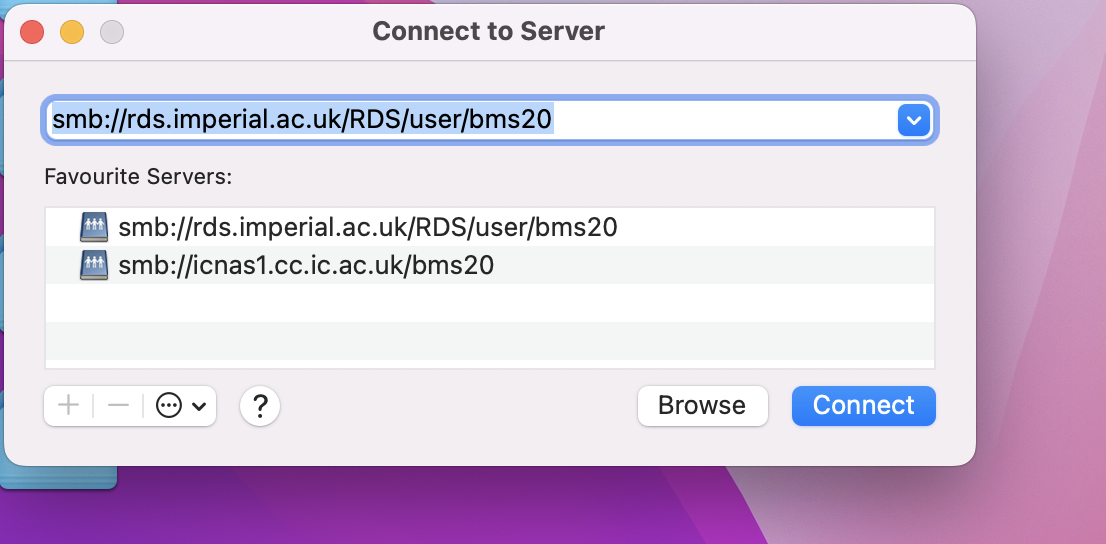
- Enter the following:
smb://rds.imperial.ac.uk/RDS/user/<YOUR_USERNAME> - Hit
ENTERand the HPC will now be mounted to your local computer atVolumes/<YOUR_USERNAME>.
You can add the following command to your ~/.bashrc so that it will be available to you whenever you open a new terminal.
To change where you want to mount the directory to, simply change the last argument:
/home/$USER/RDS --> /<YOUR PREFERRED_DIR>/$USER/RDS
alias mountrds='sudo mount -t cifs -o uid=$USER -o gid=emailonly -o username=$USER //rds.imperial.ac.uk/RDS /home/$USER/RDS'
Then, you can simply run the following to mount your HPC folder:
mountrds
If you want an R package installed and there's something that needs root permissions to setup, then request that it be installed via ASK.
You might want to try joining RCS Slack and raising the issue there.
Before doing either of these, you might want to check if the software is listed under module avail. If it is, you might be able to access it within RStudio using the RLinuxModules package. Another option is to setup the software up within conda, but that won't help with RStudio.
To create an interactive session on the cluster (to avoid overloading the login nodes) use the following command
qsub -I -l select=1:ncpus=8:mem=96gb -l walltime=08:00:00
That command requests the most resources that can be obtained for interactive jobs, decrease these if you can. On the main queue it will take a long time to submit. If I've given you access to the med-bio queue (ask) then you will be better of using the following:
qsub -I -l select=1:ncpus=2:mem=8gb -l walltime=01:00:00 -q med-bio
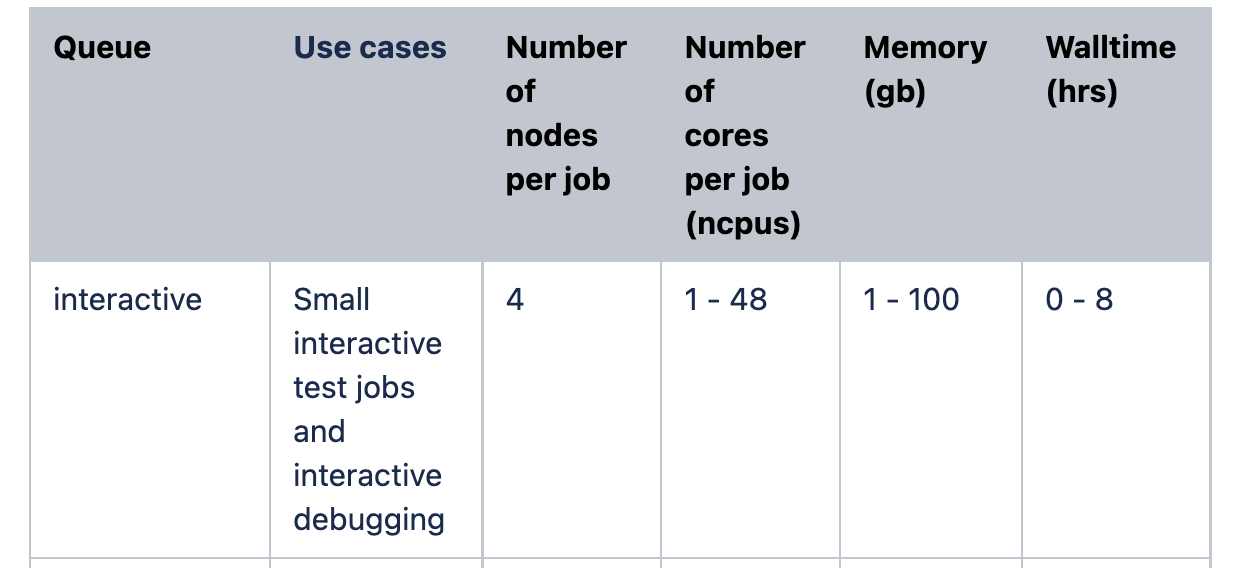
Here are the current allowed job sizing parameters for Imperials' HPC:
We have two shared project spaces on the cluster. If you are involved in the DRI Multiomics Atlas project then use projects/ukdrimultiomicsprojects/. Otherwise, please use projects/neurogenomics-lab
You will not be able to write into the main directory of either of these. They have two folders: live and ephemeral. Read about the differences between these here:
The MedBio cluster has additional computational resources and is accessed via a separate queue. Read about it here: https://www.imperial.ac.uk/bioinformatics-data-science-group/resources/uk-med-bio/
We have access to it but I do not have admin rights to grant access to individuals.
To get access it was previously necessary to email p.blakeley@imperial.ac.uk but he is no longer at Imperial, and the access management doesn't seem to have been cleared up. Another contact email is medbio-help@imperial.ac.uk. Most recently, Brian got access by emailing m.futschik@imperial.ac.uk.
Nathan Skene can access a list of users that currently have admin rights over medbio through the self-service portal. Ask him to check who is on there currently, and then contact one of them. Currently Abbas Dehghan seems like a good candidate.
To run on the med bio cluster, just put this at the end of your submit commands: -q med-bio
It can take a long time to get jobs submitted on the cluster. We can pay to get jobs submitted faster. Details are here: https://www.imperial.ac.uk/admin-services/ict/self-service/research-support/rcs/computing/express-access/. I would rather pay for faster results if this is slowing you down. Let me know if this would be useful and I'll add you to the list. Express jobs are sunmitted using Run express jobs with qsub -q express -P exp-XXXXX, substituting your express account code.
You'll need to use singularity to run docker containers on the HPC. To run a rocker R container in interactive mode, run the following, substituting your username where appropriate:
mkdir /rds/general/user/$USER/ephemeral/tmp/
singularity exec -B /rds/general/user/$USER/ephemeral/tmp/:/tmp,/rds/general/user/$USER/ephemeral/tmp/:/var/tmp,/rds/general/user/$USER/ephemeral/rtmp/:/usr/local/lib/R/site-library/ --writable-tmpfs docker://rocker/tidyverse:latest R
To create a Singularity image, first archive the image into a tar file. Obtain the IMAGE_ID with docker images then archive with (substituting the IMAGE_ID): -
docker save 409ad1cbd54c -o singlecell.tar
On a system running singularity-container (>v3) (e.g. on the HPC cluster), generate the Singularity Image File (SIF) from the local tar file with: -
/usr/bin/singularity build singlecell.sif docker-archive://singlecell.tar
This singlecell.sif Singularity Image File (SIF) is now ready to use.
On HPC, Rocker containers can be run through Singularity with a single command much like the native Docker commands, e.g.
singularity exec docker://rocker/tidyverse:latest R
By default singularity bind mounts /home/$USER, /tmp, and $PWD into your container at runtime.
More info is available here.
If you go to this link in the compute tab there is your jobs. For some jobs you can extend the walltime.
- Home
- Useful Info
- To do list for new starters
- Recommended Reading
-
Computing
- Our Private Cloud System
- Cloud Computing
- Docker
- Creating a Bioconductor package
- PBS example scripts for the Imperial HPC
- HPC Issues list
- Nextflow
- Analysing TIP-seq data with the nf-core/cutandrun pipeline
- Shared tools on Imperial HPC
- VSCode
- Working with Google Cloud Platform
- Retrieving raw sequence data from the SRA
- Submitting read data to the European Nucleotide Archive
- R markdown
- Lab software
- Genetics
- Reproducibility
- The Lab Website
- Experimental
- Lab resources
- Administrative stuff