-
Notifications
You must be signed in to change notification settings - Fork 789
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Custom factor (unaryFactor) issue with initial heading #704
Comments
|
Did you init test your derivatives? Numerous examples to do this with numerical differentiation. Or use the super-handy |
|
For this test I used the original UnaryFactor class from the LocalizationExample.cpp The Jacobian is definitely correct for the given error function, however the question is if the formulated error function is valid. As shown by the results of the test case, the optimizer cannot converge to the solutions... |
|
I'm not so convinced about the Jacobian. How about quickly calling EXPECT_CORRECT_FACTOR_JACOBIANS now that you have everything set up? We might be surprised, as Jacobian is about the tangent space on the right. |
|
I didn't manage to get the 1. No orientation: result: this is ok since testFactorJacobians returns 2. 90° orientation:
|
|
It seems that the expected Jacobian is as follows: |
|
Indeed, this is what I suspected :-) Try correcting the factor and see whether everything works? If so, a PR with the change would be much appreciated. |
|
I tested the factor with correct Jacobian and it seems to be working perfect with real data as well as synthetic ones. I will send a PR, but for my understanding gtsam expect to have error and Jacobian in tangent space right? |
|
This is the PR for fixing the Jacobian: I think as an improvement, we can have UnaryFactor class as a stand-alone factor. |
Description
It seems that when using a custom factor for GPS measurements (UnaryFactor class as in the instructions) with a prior node with initial heading, the solvers in C++ cannot optimize and converge to the solutions. Setting the heading to 0° however poses no issue and the solvers can converge after some iterations even with noisy initial estimates. This has been tested both with synthetic data and real data and both give similar results but with the real data it is even worst.
Steps to reproduce
-------------------------------------------------------------------If we extend LocalizationExample.cpp by adding a prior factor (
PriorFactor) with a 90° heading as below and also updating the GPS data and initial estimates, then neither of the solvers (LevenbergMarquardtOptimizer / GaussNewtonOptimizer) can solve/converge.LevenbergMarquardtOptimizerthrows the following for instance:-------------------------------------------------------------------Here is the code I tested with, compared to LocalizationExample.cpp, a prior node is added and also
initialEstimateandUnaryFactorare updated.-------------------------------------------------------------------Here is the print out when setting the verbosity to
"ERROR":Expected behavior
gtsam should converge to the GPS positions as shown in the below figure plotted by Matlab gtsam-toolbox-3.2.0-win64:
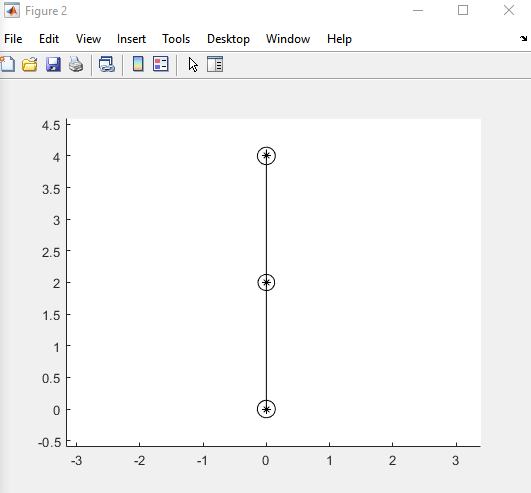
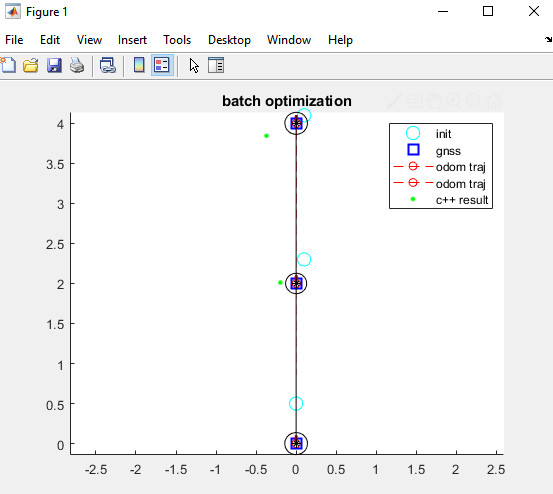
However the c++ solutions look like the green dots in the following picture:
Environment
We have used C++ as the language together with gtsam 4.0.3 in Ubuntu 18.04. Matlab gtsam-toolbox-3.2.0-win64 is used for simulations and plotting. Note that in Matlab
PriorFactorPose2is used as the equivalent for theUnaryFactorin C++ since we did not figured how to add a custom factor in Matlab. So the comparison of the results between Matlab and C++ is not entirely fair but C++ is expected to give decent results.Additional information
The text was updated successfully, but these errors were encountered: