-
Notifications
You must be signed in to change notification settings - Fork 6
Containerized Pipelines
A 'Pipeline' is defined as an automated sequence of data processing steps where the output of the first step becomes the input of the next step, etc. Throughout this course we have presented many python scripts to automate geospatial data processing. A script with several sequential steps could be considered a 'pipeline'.

For the purposes of this lesson we are interested in connecting sequential processing steps between multiple types of software. For example, we may want to use a combination of command line tools, Python, Javascript, and R to go from the first to the last step of our pipeline. Fortunately, there are several software options to link multiple disparate software together into a sequential pipeline. This gives us the power to automate and combine our favorite software even if they are from different ecosystems.

This lesson is going to demonstrate a small geospatial pipeline using Docker-Compose. Other pipeline or workflow management software out there include Nextflow, Makeflow, and Snakemake.
Sharing your scientific analysis code with your colleagues is an act of collaboration that will help push your field forward. There are, however, technical challenges that may prevent your colleagues from effectively running the code on their own computer. These include:
- hardware: CPUs, GPUs, RAM
- Operating System: Linux, MacOS, Windows
- Software version: R, Python, etc
- Library versions and dependencies
How do we make it easier to share analysis code and avoid the challenges of computer and environment setups?
A container is a standard unit of software that packages up code and all its dependencies so the application runs quickly and reliably from one computing environment to another. Container images are a lightweight, standalone, executable package of software that includes everything needed to run an application: code, runtime, system tools, system libraries and settings. Each of these elements are specifically versioned and do not change. The user does not need to install the software in the traditional sense.
A useful analogy is to think of software containers as shipping containers. It allows us move cargo (software) around the world in standard way. The shipping container can be offloading and executed anywhere, as long the destination has a shipping port (i.e., Docker).

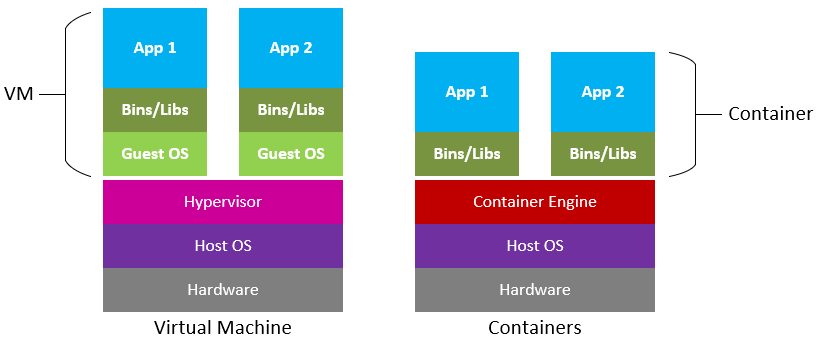
Containers are similar to virtual machines (VMs), but are smaller and easier to share. A big distinction between Containers and VMs is what is within each environment: VMs require the OS to be present within the image, whilst containers rely on the host OS and the container engine (e.g., Docker Engine).
Software containers, such as those managed by Docker or Singularity, are incredibly useful for reproducible science for several reasons:
Containers encapsulate the software environment, ensuring that the same versions of software, libraries, and dependencies are used every time, reducing the "it works on my machine" problem.
Containers can be easily shared with other researchers, allowing them to replicate the exact software environment used in a study.
Containers can run on different operating systems and cloud platforms, allowing for consistency across different hardware and infrastructure.
The most common container software is Docker, which is a platform for developers and sysadmins to develop, deploy, and run applications with containers. Apptainer (formerly, Singularity), is another popular container engine, which allows you to deploy containers on HPC clusters.
DockerHub is the world's largest respository of container images. Think of it as the 'Github' of container images. It facilitates collaboration amongst developers and allows you to share your container images with the world. Dockerhub allows users to maintain different versions of container images.
Now that you have a basic understanding of Pipelines and Containers, let's combine the two. We are going to do a hands-on exercise where we run a series of containerized software in a processing pipeline. For our example, we will be processing drone imagery that was collected over a golf course in Phoenix, AZ.

-
Input data are 32 jpeg images collected from a drone flying at ~60 m above the ground. The images are overlapping and include nadir and oblique angles. The camera was a 20 megapixels RGB sensor from a DJI Phantom 4 RTK drone.
-
The first software container is OpenDroneMap. ODM is a command-line open-source photogrammetry software package that takes a series of overlapping aerial images and creates imagery products such as 3D point clouds, digital elevation models, and orthomosaics. In our pipeline, the output of ODM is an unclassified point cloud.
-
The second container is another command-line tool called PDAL. It is useful for all kinds of point cloud analysis. In our pipeline, the PDAL will take the output from ODM (an unclassified point cloud), and execute a classification procedure (i.e, filtering) to identify points that represent vegetation and points that represent ground. The specific filtering method used is called Cloth Simulation Filtering.
-
Each of these steps is an individual docker container. They are orchestrated and ran sequentially using a docker-compose yml configuration file.
The instructions for running the pipeline are found in the following Github repository: https://github.com/jeffgillan/geospatial_pipeline
