-
Notifications
You must be signed in to change notification settings - Fork 403
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Include 21L rooted Nextstrain build for GISAID data #1029
Conversation
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
I've noticed that you use a 21L reference with deletions.
Will we keep using Wuhan coordinates? This should be the case as long as we translate using Wuhan - otherwise Spike and other positions will be messed up.
|
I think best to keep everything in Wuhan/1 coordinates. Latest BQ.1 preprints still use Wuhan/1 coordinates. Will be way too confusing otherwise. So perhaps this PR is better thought of as 21L "root" rather than 21L "reference". |
|
I totally agree, especially given that deletions commonly occur, like 69/70del and 144del etc it'd be a mess to change coordinate systems and not worth it. |
|
Commenting mostly on the organizational/conceptual bits (and not the code changes here)…
Why not keep existing builds (with the ancestral Wuhan root) at i.e. don't change what's already there, only add the
Alternatively, a pre-subsampling filter rule could be added to the DAG for this profile instead of moving the existing inputs:
- name: gisaid
metadata: "results/gisaid_21L_metadata.fasta.xz"
aligned: "results/gisaid_21L_aligned.fasta.xz"a new custom_rules:
- nextstrain_profiles/nextstrain-gisaid-21L/prefilter.smkand in that new file writing the rule to do the filtering down to 21L, e.g. something that looks roughly like: rule 21L_prefilter:
input:
metadata = path_or_file("s3://nextstrain-ncov-private/metadata.tsv.xz"),
aligned = path_or_file("s3://nextstrain-ncov-private/aligned.fasta.xz")
output:
metadata = "results/gisaid_21L_metadata.tsv.xz",
aligned = "results/gisaid_21L_aligned.fasta.xz"
shell:
…(You also might be able to accomplish the same thing without any custom rules by chaining inputs together, though I'm not sure it's possible with existing config.) I don't think this as a one-off approach is necessarily better than one-off provisioning of a separate set of files upstream in ncov-ingest, but I think if the prefiltering concept was generalized for this workflow (even if prefiltering was still limited only to clades or something) that might be better than producing/maintaining lots of separate static subsets.
Is the biggest annoyance here the slowness of
I think it's reasonable to provision separate subsets for specific cases like this, though I do wonder if the alternative I describe above might be better in the long run. Re: these specific proposed paths: they do perpetuate the comingling of workflow-associated files and dataset-associated files that ncov-ingest and ncov already do, and in that sense it's fine and maybe even preferred, but it might be nice to consider putting new workflow-associated files (which these are) in the new locations we decided upon, e.g.: Since we're not planning to remove the existing comingled files for ncov/ncov-ingest, avoiding adding more might help avoid further confusion of what's what. |
| Adjusting metadata for build '{wildcards.build_name}' | ||
| """ | ||
| input: | ||
| metadata="results/{build_name}/metadata_with_index.tsv", | ||
| metadata="results/{build_name}/metadata_filtered.tsv.xz", | ||
| output: | ||
| metadata = "results/{build_name}/metadata_adjusted.tsv.xz" |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
This changes the metadata that's input to several downstream rules. Rules which take metadata_adjusted.tsv.xz as input currently get the subsampled but unfiltered metadata and with this change would get the subsampled and filtered metadata.
Is this change a bug fix? It seems like it. But it does have potential visible changes like rule colors now will produce a scale based on the filtered rather than unfiltered metadata values.
I'd appreciate someone else's 👀 on this bit.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
/cc @nextstrain/core
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Split out separately in d8c17ec.
|
Rebased this onto the latest master to bring in changes. This also highlights a few things that need updating since the copy, which I'll mark separately with comments here so I can track them. |
nextstrain_profiles/nextstrain-gisaid-21L/nextstrain_description.md
Outdated
Show resolved
Hide resolved
72992da to
bb384a7
Compare
This is intended as a bug fix to more closely align metadata with sequences in rules downstream of subsample + filter. Rules which take metadata_adjusted.tsv.xz as input used to get the subsampled _but not_ filtered metadata. With this change, they now get the subsampled _and_ filtered metadata. Many rules will not be affected as they join sequences → metadata and ignore extra metadata rows. However, there are some potential changes to results as some rules do consult all of the metadata without joining it to sequences. For example, the "colors" rule will now produce a scale based on the filtered rather than unfiltered metadata values, which could be noticeably different sets. Co-authored-by: Trevor Bedford <trevor@bedford.io>
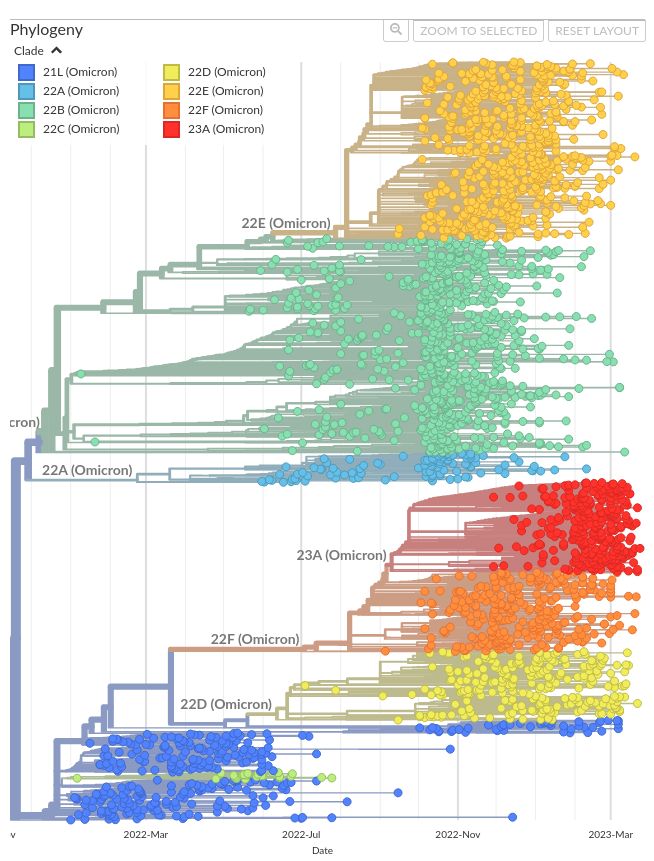
All circulating viruses now descend from clade 21L. By rooting the tree on 21L reference virus we can better emphasize recent evolution. This includes tree layout, but also things like S1 mutations, where these were basically all just red rather than emphasizing differences among circulating viruses, or clades, where all the previous clades were making 21L through 22E all similar colors of red. This copies the existing Nextstrain GISAID build and adds some simple logic to filter out clades previous to 21L. This results in some code duplication between the ancestral GISAID build and the 21L GISAID build, but overall much simpler code than if a single build.yml covered both ancestral and 21L root cases. Co-authored-by: Thomas Sibley <tsibley@fredhutch.org>
Filter rules in the config are applied _after_ subsampling, which poses issues with reliably getting the desired number of sequences. As @trvrb wrote¹: > In the workflow, the filter rule happens after the subsampling rules. > This makes it so that if we ask for say 2560 in a sampling bucket, we'll > lose >50% due to filtering out non-21L-descending clades. > > This could be solved by padding count targets to compensate, but this is > hacky and the numbers will change as time goes on. Or the filter rule > could be placed again before subsample, but we moved it afterwards for > good reasons. A few custom rules for the builds allows us to prefilter the full dataset before subsampling. Currently these rules are specific to our GISAID data source, but they could be easily expanded to our Open data sources too. In the future we might also provide clade-partitioned subsets from ncov-ingest², which we could use here instead with some adaptation of the build config. ¹ <#1029 (comment)> ² e.g. <nextstrain/ncov-ingest#398>
|
Per discussion last week, I reworked this branch to prefilter the data before subsampling and to avoid memory-inefficient steps. See the commit messages for details. This is ready for review again by @nextstrain/core (and maybe particularly @trvrb). There's also a trial build running now. |
|
Looking at the global/6m build locally, it seems ok. Oddly, I noticed there's a sequence that made it thru classified as 19A: This sequence is in our metadata as 22B, so passes the pre-filters, and is from 2022-12, so passes the date filter, but must be getting reclassified to 19A (erroneously?) by the workflow. I haven't dug into it more, but perhaps we want to both pre-filter and (post-)filter on the clades to catch these reclassifications? |
|
Yes - in fact, I bet it's not even the workflow proper where the reclassification is happening - it's just at the end when the clades are being 'put' on the tree (so that the labels show up and everything gets a nice colour). This overrides whatever classifications we've gotten from Nextclade in the display. But these are essentially only assigned at one node - then everything downstream from that is classified as "being that node" (unless another node downstream also has a label, which then overrides the first). If I'm remembering correctly, node labels attach to the first node available that matches their specified mutations. 19A is a bit 'weak' in the respect that it's only defined by 2 mutations -- of course, we didn't have many muts at the start!! So this may well literally be all that defines this clade. Naively I would have said that node assignments happen in order specified in the TSV, with nodes coming later in the list overriding any assigned earlier (after all, after the first clade is assigned 1/2 the tree may be 'part of that node' until you do your next assignment within that) - so I'm a little surprised this happened, but I may be completely misremembering how this works. If useful for others, here's the tree: https://nextstrain.org/staging/ncov/gisaid/21L/trial/21L-reference/global/6m?label=clade:22E%20%28Omicron%29&showBranchLabels=all |
|
As for solutions: I don't think a pre-filter will help, as this node really is 22B, it's just that it also has these two 19A mutations. The most straightforward (on the surface) solution would be to use a separate clades_nextstrain.tsv for these 21L builds - but already have a lot of these and we have to update each separately every time we add a new clade - so that's not ideal if we can find another way... |
Ahh, this makes sense. Thanks for the explanation!
Hmm. I'd think we could programmatically filter the clades though to remove the ones prior to 21L… I'll try it. |
|
Oh, hrm, but I can't just remove the excluded clades because they're defined hierarchically. So the filtering/transformation is a bit more complex. |
|
I've pushed 59a672c to try to address the clade labeling issue. Testing a global/6m build first, then will trigger a full set of trial builds. |
|
Very nice little solution @tsibley ! I like the idea and the implementation - makes sense to me, and avoid us having another clades file to update manually! |
This avoids incorrect labeling of the trees by `augur clades` if a sequence happens to have mutations that define one of the excluded clades. This happens because there aren't earlier sequences for those clades to be labeled due to their exclusion. For example, we've seen some spurious labelings of 19A because we exclude such sequences from these builds. Thanks to @emmahodcroft for debugging and explaining this!¹ Since we define our clades hierarchically with inheritance, a small new program is needed to fully expand those definitions so we can filter them without worrying about filtering out their "parent" clades. ¹ <#1029 (comment)>
|
Off-by-one error on my part because I saw but then didn't recall this bit about
|
|
I've started another full set of trial builds (build id |
|
Looks like the test builds completed, and checking a few of them, they look really good now! Thanks for tackling this Tom, I think it'll be really useful! |
|
LGTM, but just a heads up that the But for the time being this should work, just highlighting possible future divergence and breakage. |
|
@rneher Yep, thanks for noting that. I think the sort of thing done by this PR is a great example of why we want to do nextstrain/ncov-ingest#399. :-) I'd much rather write and maintain code to filter on |
This should have been removed in "Remove preprocessing pipelines" (9abad2a).
This is the event triggered by ncov-ingest when there's new GISAID data, which also triggers the standard ancestral-rooted builds. The specific "rebuild-gisaid-21L" event for this workflow seems unnecessary since it would only be triggered manually, and in that case I expect folks to use the GitHub Actions web UI instead.
|
Pushed a couple additional small commits, one of which ensures these 21L-rooted builds are triggered by new data, and now will merge. |
|
One thing I think that's worth addressing, but that I chose not to address now (because I have to timebox this to some reasonable scope), is the vast duplication between |
|
Five months ago, I wrote:
The above is what I've done in getting this PR over the line in the past two weeks. One reason, given on the biweekly call right now by @trvrb, to switch existing builds to use an ancestral label, is that it would make it possible to update nextstrain.org's dataset manifest (ugh that this still exists) such that the dataset selector dropdowns work when viewing 21L builds (and 21L builds are navigable from the ancestral builds). Currently the dataset selector dropdowns do not work well for 21L builds. I'd suggest that instead of continuing to shift existing URLs all the time and maintain all those redirects, we could instead prioritize a better dataset selector (such as the one sketched out by @jameshadfield) and subsequent removal of the manifest. |
|
Good point about dataset selector. We currently also have this issue for https://nextstrain.org/nextclade/sars-cov-2/21L. |
|
Also, I think we can get okay, but not ideal, behavior in the dataset selector from the current dataset names of:
by modifying The issue with https://nextstrain.org/nextclade/sars-cov-2/21L is having |
|
BTW I definitely didn't intend to sound 'problem-finding' with the drop-downs - apologies if it came across that way! I think it's fantastic the 21L build is up, and thanks for pushing it over the line, @tsibley! I only brought it up because the drop-down is mainly how I remember what builds we have and how to get to them (so I find it really useful). But, I totally get the issue at play here, and I agree that constantly modifying URLs is not an ideal solution. Coming up with a "better way" is almost certainly going to pay off in the long-run so we can have whatever URLs we want, without needing 'sisters' etc. |
|
@emmahodcroft No worries! I have no problem (ha) with you pointing out the dropdown problems—they're real obstacles to usage!—I just wanted to make it clear here that a) the problem exists (since it hadn't been written down) but that b) I'm personally not going to solve it at this time. :-) |



Description of proposed changes
This PR includes a new set of Nextstrain builds for GISAID data that use 21L as reference rather than Wuhan. All viruses that outgroup relative to 21L in the tree are dropped via
filterrule:I decided to split out a separate
nextstrain_profiles/nextstrain-gisaid-21Lrather than padding outnextstrain_profiles/nextstrain-gisaidwith additional logic because the latter would have resulted in extensive code-duplication within thebuilds.yamlin specifying different behavior across specific build endpoints.I've run this locally with swapping
to
and things work well and run quickly. I'm still testing using the full data on AWS however and will update this PR with links to trial runs.
That said, restricting to 21L makes a number of visuals much clearer. For example, here is the smaller local tree looking at S1 mutations:
I'd plan to split things out so that available endpoints include:
nextstrain.org/ncov/gisaid/ancestral/global/6mnextstrain.org/ncov/gisaid/21L/global/6mnextstrain.org/ncov/gisaid/21L/asia/2metc...
However, all this said, I don't think this is the right tact to solve the problem.
Issues include:
In the workflow, the
filterrule happens after the subsampling rules. This makes it so that if we ask for say 2560 in a sampling bucket, we'll lose >50% due to filtering out non-21L-descending clades.This could be solved by padding count targets to compensate, but this is hacky and the numbers will change as time goes on. Or the
filterrule could be placed again beforesubsample, but we moved it afterwards for good reasons.I'd anticipate wanting a number of analyses (like https://github.com/blab/ncov-escape and other projects) that use 21L as reference. Having to deal with the entire dataset and having to filter out outgroup viruses will be repeatedly annoying.
Instead, I'd propose that we move this
filterlogic to https://github.com/nextstrain/ncov-ingest and to make the filtered metadata and sequences available for both GISAID data and open data at URLs like:s3://nextstrain-ncov-private/21L/metadata.tsv.gzs3://nextstrain-ncov-private/21L/sequences.fasta.xzs3://nextstrain-data/files/ncov/open/21L/metadata.tsv.gzs3://nextstrain-data/files/ncov/open/21L/sequences.fasta.xzThis will streamline and speed up running of 21L builds (
augur filterruns repeatedly on a smaller metadata file) and help enable many future analyses.Testing
As above, I've locally tested with
and trial build is running that will output to https://nextstrain.org/staging/ncov/gisaid/21L/trial/21L-reference/global/6m. I'll update when this completes.