aka Generic Optimization by Genetically Evolved Operation Trees
This library allows feature engineering by genetically evolved operation trees
(in fact they are DAGs, but geopd sounded ugly).
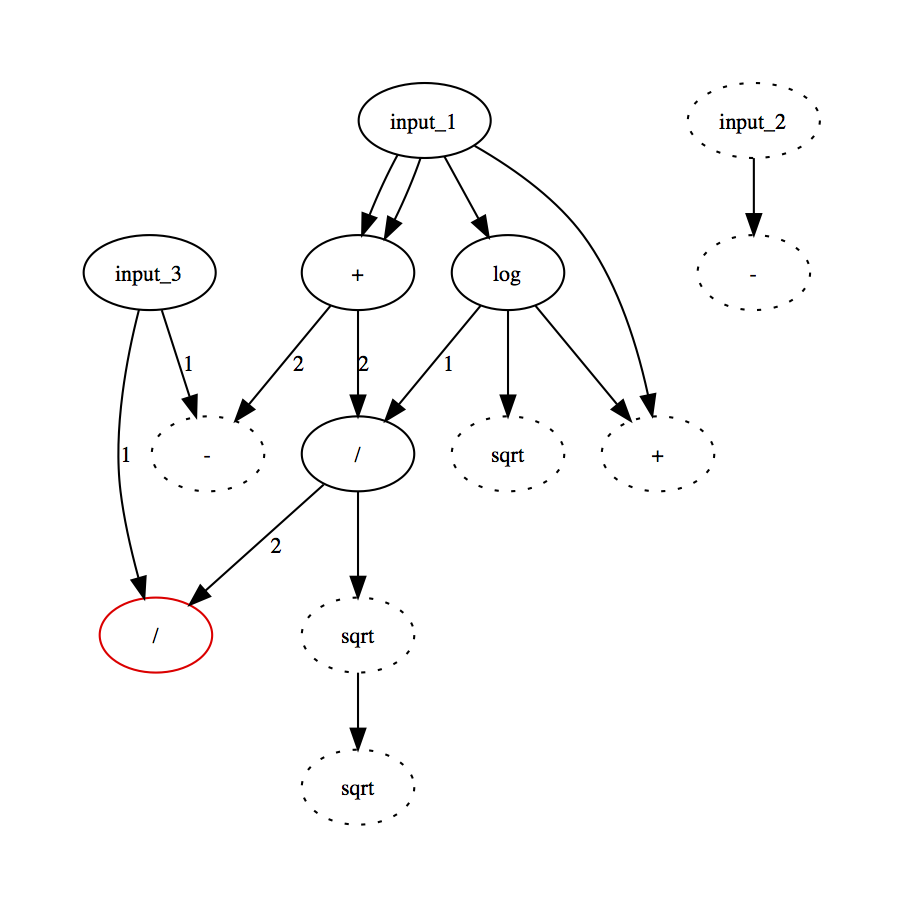
The trees look like this (output in red):
The most two most important optimizations are:
- the reduction of the operation trees by using SymPy
- good numerical performances when using the numexpr backend (optional argument of
OpTree.evaluate).
The Evolution.evolve can also take a pool argument and be parallelized.
The binary_classification.ipynb notebook shows how a simple threshold model
trained on 20% of the data yields 95.6% accuracy on the
Breast Cancer Wisconsin (Diagnostic) Data Set
The examples include:
binary_classification.ipynb: simple binary classification with threshold evolved with area under ROC curvesymbolic_regression.ipynb
geopt can be used as the first layer of any model. For example it could be of great help
as feature engineering in linear classifiers or neural networks.
However, you want the model used in the fit function to be quite cheap to evaluate,
even if you use another model in the end.
This is just a proof of concept, the pull requests are very welcome.
Look at the examples/ folder.
Also, the code is really not big (~ 600 SLOC), quite readable and well commented.
- Add numeric parameter nodes, compute the derivatives using SymPy and do gradient descent
- Since many OpTrees are similar in the population and over the generations,
one could memoize their fitness on the current dataset. A simple way to do it would be to make a dictionnary
{formula: fitness}. - One could think this one step further with a node grained memoization
{node: evaluation}, with the drawback that it could only work with the 'raw' numpy backend (backend=None). However, the memory footprint would be horrible, and numexpr offers great performances so I don't think it would be a good idea. - Clustering would be possible since the OpTrees are picklable, with an interface like this root client and those node servers.