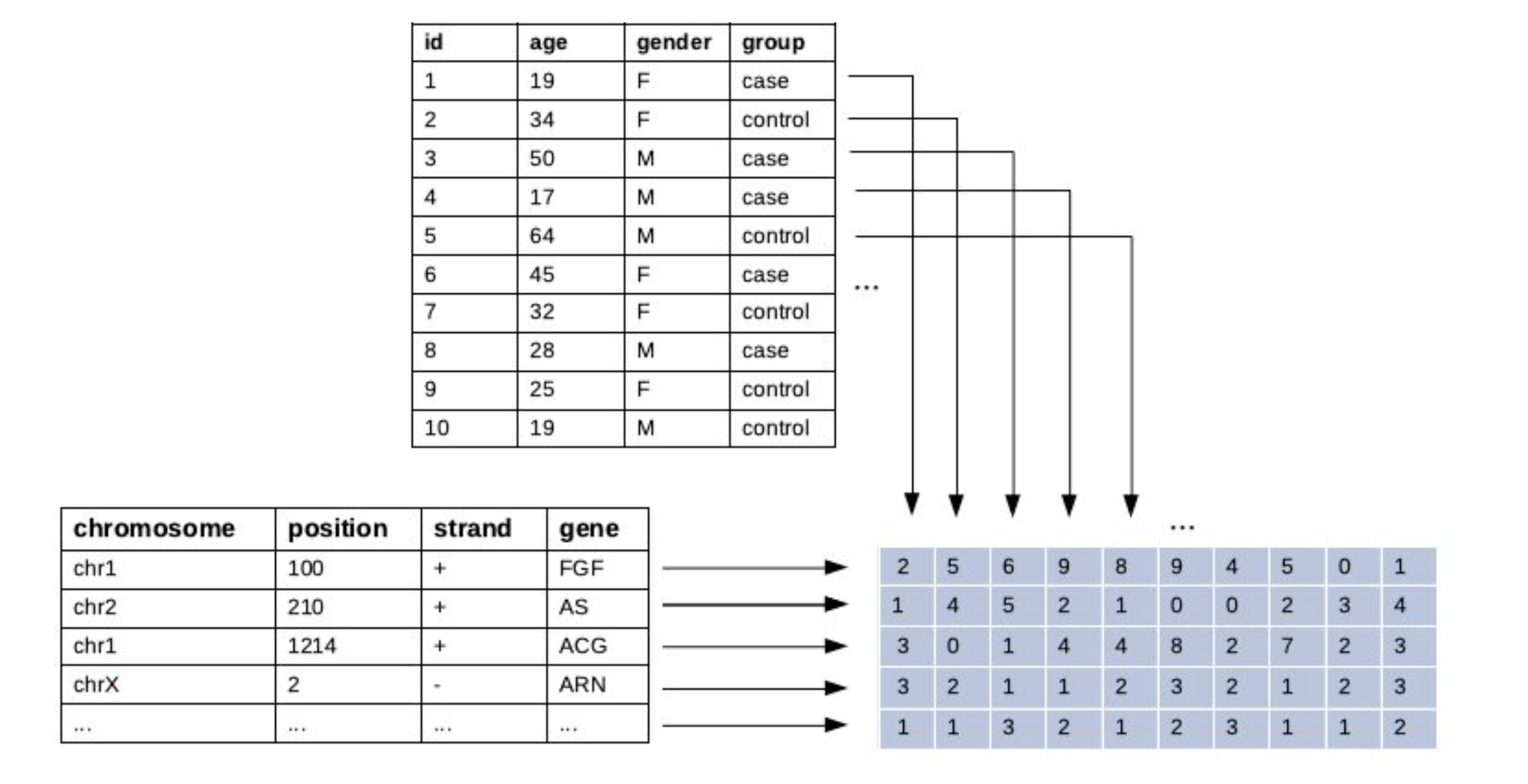
R Annotated Matrix Object
annmatrix implements persistent row and column annotations for R matrices.
The use-case was born out of the need to better organize biomedical microarray and sequencing data within R. But 'annmatrix' is readily applicable in other contexts where the data can be assembled into a matrix form with rows and columns representing distinct types of information.
The main advantage of 'annmatrix' over BioConductor implementations like SummarizedExperiment and AnnotatedDataFrame is simplicity. Since 'annmatrix' is based on a matrix, and not a list or a data frame, it behaves like a regular matrix and can be directly passed to various methods that expect a matrix for an input.
Say, we have a small gene expression dataset with 10 genes measured across 6 samples.
mat <- matrix(rnorm(10 * 6), nrow = 10, ncol = 6)
[,1] [,2] [,3] [,4] [,5] [,6]
[1,] -0.66184983 -0.3828219 -1.2668148 -1.42199245 -0.431174368 -1.86544873
[2,] 1.71895416 0.2994216 -0.1985833 -0.32822829 0.382446538 1.82998433
[3,] 2.12166699 0.6742398 0.1388658 0.28457007 0.041125095 -0.99111590
[4,] 1.49715368 -0.2928163 -0.2793360 0.71933588 -0.059224001 -1.45043462
[5,] -0.03614058 0.4880534 0.7089194 0.43241598 -1.296423984 -0.01482476
[6,] 1.23194518 0.8828018 -0.7666105 -0.35192477 -2.136976377 0.53663905
[7,] -0.06488077 1.8627490 1.4433629 0.29772143 -0.893614686 -0.81110377
[8,] 1.06899373 1.6117253 0.8448793 -0.26143236 0.612732875 -0.31832480
[9,] -0.37696531 0.1354795 -0.3993704 1.30868973 0.582971232 1.11147600
[10,] 1.04318309 1.0880860 -1.4277676 0.01587026 -0.005882013 -0.14174471We can turn this matrix into 'annmatrix' object right away.
X <- as.annmatrix(mat)When printed annmatrix shows 4 first + the last row and 4 first + the last column from the matrix and lists all available row and column annotations (of which there are currently none).
X
[,1] [,2] [,3] [,4] [,6]
[1,] -0.66184983 -0.38282188 -1.26681476 -1.42199245 ........... -1.86544873
[2,] 1.71895416 0.29942160 -0.19858329 -0.32822829 ........... 1.82998433
[3,] 2.12166699 0.67423976 0.13886578 0.28457007 ........... -0.99111590
[4,] 1.49715368 -0.29281632 -0.27933600 0.71933588 ........... -1.45043462
........... ........... ........... ........... ........... ...........
[10,] 1.04318309 1.08808601 -1.42776759 0.01587026 ........... -0.14174471
rann:
cann:Custom operators @ and $ are provided for convenient manipulation of row and column data.
Let's add some additional information about our samples.
X$group <- c("case", "case", "case", "control", "control", "control")
X$sex <- c("F", "M", "M", "M", "F", "F")
X
[,1] [,2] [,3] [,4] [,6]
[1,] -0.66184983 -0.38282188 -1.26681476 -1.42199245 ........... -1.86544873
[2,] 1.71895416 0.29942160 -0.19858329 -0.32822829 ........... 1.82998433
[3,] 2.12166699 0.67423976 0.13886578 0.28457007 ........... -0.99111590
[4,] 1.49715368 -0.29281632 -0.27933600 0.71933588 ........... -1.45043462
........... ........... ........... ........... ........... ...........
[10,] 1.04318309 1.08808601 -1.42776759 0.01587026 ........... -0.14174471
rann:
cann: group, sex
We can add the entire annotation table by assigning to an empty value. Let's do this for rows.
chromosome <- c("chr1", "chr1", "chr1", "chr1", "chr2", "chr2", "chr2", "chr3", "chr3", "chr3")
position <- runif(10, 0, 1000000)
X@'' <- data.frame(chr = chromosome, pos = position)
X
[,1] [,2] [,3] [,4] [,6]
[1,] -0.66184983 -0.38282188 -1.26681476 -1.42199245 ........... -1.86544873
[2,] 1.71895416 0.29942160 -0.19858329 -0.32822829 ........... 1.82998433
[3,] 2.12166699 0.67423976 0.13886578 0.28457007 ........... -0.99111590
[4,] 1.49715368 -0.29281632 -0.27933600 0.71933588 ........... -1.45043462
........... ........... ........... ........... ........... ...........
[10,] 1.04318309 1.08808601 -1.42776759 0.01587026 ........... -0.14174471
rann: chr, pos
cann: group, sexSame shortcuts can be used for retrieving available annotations.
X@chr
[1] "chr1" "chr1" "chr1" "chr1" "chr2" "chr2" "chr2" "chr3" "chr3" "chr3"
X$group
[1] "case" "case" "case" "control" "control" "control"As well as for adjusting annotations.
X@pos
[1] 418638.0 423928.7 248000.4 220840.4 482854.1 662214.0 122708.6 334171.7 219852.3 927672.1
X@pos <- X@pos * 10
X@pos
[1] 4186380 4239287 2480004 2208404 4828541 6622140 1227086 3341717 2198523 9276721Or create new ones.
X$age <- seq(10, 60, 10)
X$age
[1] 10 20 30 40 50 60When an empty name is provided it will return the whole annotation data.frame.
X$''
group sex age
1 case F 10
2 case M 20
3 case M 30
4 control M 40
5 control F 50
6 control F 60When subsetting the annmatrix object all the annotations are correctly adjusted and class is preserved.
X_case <- X[, X$group == "case"]
X_case
[,1] [,2] [,3]
[1,] -0.6618498 -0.3828219 -1.2668148
[2,] 1.7189542 0.2994216 -0.1985833
[3,] 2.1216670 0.6742398 0.1388658
[4,] 1.4971537 -0.2928163 -0.2793360
.......... .......... ..........
[10,] 1.0431831 1.0880860 -1.4277676
rann: chr, pos
cann: group, sex, ageX_case$''
group sex age
1 case F 10
2 case M 20
3 case M 30However in order to be consistent with matrix the class is dropped when selecting only a single row or column.
X[1,]
[1] -0.6618498 -0.3828219 -1.2668148 -1.4219925 -0.4311744 -1.8654487But just like with a matrix we can enforce it to preserve all the annotations and the class by setting drop=FALSE.
X[1,, drop=FALSE]
[,1] [,2] [,3] [,4] [,6]
[1,] -0.6618498 -0.3828219 -1.2668148 -1.4219925 .......... -1.8654487
rann: chr, pos
cann: group, sex, ageOperations on annmatrix object don't loose the class.
X > 0
[,1] [,2] [,3] [,4] [,6]
[1,] FALSE FALSE FALSE FALSE ..... FALSE
[2,] TRUE TRUE FALSE FALSE ..... TRUE
[3,] TRUE TRUE TRUE TRUE ..... FALSE
[4,] TRUE FALSE FALSE TRUE ..... FALSE
..... ..... ..... ..... ..... .....
[10,] TRUE TRUE FALSE TRUE ..... FALSE
rann: chr, pos
cann: group, sex, ageX <- X - rowMeans(X)
X
[,1] [,2] [,3] [,4] [,6]
[1,] 0.34316717 0.62219512 -0.26179776 -0.41697545 ........... -0.86043172
[2,] 1.10162165 -0.31791091 -0.81591579 -0.94556080 ........... 1.21265182
[3,] 1.74344169 0.29601446 -0.23935952 -0.09365523 ........... -1.36934120
[4,] 1.47470725 -0.31526276 -0.30178244 0.69688945 ........... -1.47288106
........... ........... ........... ........... ........... ...........
[10,] 0.94789225 0.99279517 -1.52305843 -0.07942058 ........... -0.23703556
rann: chr, pos
cann: group, sex, ageMatrix transpose will preserve the class and correctly adjust row and column annotations.
t(X)
[,1] [,2] [,3] [,4] [,10]
[1,] 0.34316717 1.10162165 1.74344169 1.47470725 ........... 0.94789225
[2,] 0.62219512 -0.31791091 0.29601446 -0.31526276 ........... 0.99279517
[3,] -0.26179776 -0.81591579 -0.23935952 -0.30178244 ........... -1.52305843
[4,] -0.41697545 -0.94556080 -0.09365523 0.69688945 ........... -0.07942058
........... ........... ........... ........... ........... ...........
[6,] -0.86043172 1.21265182 -1.36934120 -1.47288106 ........... -0.23703556
rann: group, sex, age
cann: chr, posPrincipal component analysis with prcomp will transform the PCA loadings into 'annmatrix' object and add row and column annotations to it, such as proportion of variance explained.
pca <- prcomp(t(X))
pca$rotation
PC1 PC2 PC3 PC4 PC6
[1,] 0.17186228 -0.06111446 0.28479933 0.34143265 ........... 0.06141574
[2,] 0.07770596 0.56511346 -0.15143694 0.26206867 ........... -0.05690171
[3,] 0.48808049 0.04486714 0.31834335 -0.16657847 ........... 0.57896252
[4,] 0.34355333 0.02691052 0.45134983 -0.56375589 ........... -0.52045090
........... ........... ........... ........... ........... ...........
[10,] 0.32535557 0.32483324 0.07792832 0.15417554 ........... 0.21093439
rann: chr, pos
cann: pc, sd, var, var_explained
pca$rotation$var_explained
[1] 3.976151e-01 2.492482e-01 1.991826e-01 8.899235e-02 6.496171e-02 3.582514e-33Furthermore, matrix cross-product will preserve all annotations that are possible to preserve after the product.
X_scores <- t(pca$rotation) %*% X
X_scores
[,1] [,2] [,3] [,4] [,6]
PC1 2.642132e+00 1.845104e+00 -3.957560e-01 -5.252007e-01 ............. -1.828050e+00
PC2 1.486939e+00 -8.303449e-01 -2.172757e+00 -3.843922e-01 ............. 1.723213e+00
PC3 7.210412e-01 -8.182090e-01 -4.038344e-01 1.336113e-01 ............. -1.709567e+00
PC4 -2.734391e-01 8.671400e-01 2.275004e-03 -1.559362e+00 ............. 2.337191e-01
............. ............. ............. ............. ............. .............
PC6 1.110223e-16 2.220446e-15 2.553513e-15 -1.020364e-14 ............. -2.567391e-16
rann: pc, sd, var, var_explained
cann: group, sex, age
X_scores@''
pc sd var var_explained
PC1 PC1 1.853695e+00 3.436186e+00 3.976151e-01
PC2 PC2 1.467652e+00 2.154001e+00 2.492482e-01
PC3 PC3 1.311996e+00 1.721335e+00 1.991826e-01
PC4 PC4 8.769670e-01 7.690712e-01 8.899235e-02
PC5 PC5 7.492653e-01 5.613986e-01 6.496171e-02
PC6 PC6 1.759547e-16 3.096006e-32 3.582514e-33
X_scores$''
group sex age
1 case F 10
2 case M 20
3 case M 30
4 control M 40
5 control F 50
6 control F 60And, of course, we get all the goodies that come from storing our data as a matrix.
# medians of all genes on chromosome 1
library(matrixStats)
rowMedians(X[X@chr == "chr1",])
[1] 0.04068471 -0.27639844 -0.16650737 -0.19172644
# Gene-wise Bartlett's test for equal variance between cases and control
library(matrixTests)
row_bartlett(X, X$group)
obs.tot obs.groups var.pooled df statistic pvalue
1 6 2 0.3717333 1 0.363202329 0.546733101
2 6 2 1.0998615 1 0.016042378 0.899210686
3 6 2 0.7554444 1 0.268153616 0.604573040
4 6 2 1.1341347 1 0.006848371 0.934046441
5 6 2 0.4758403 1 1.043799502 0.306939452
6 6 2 1.4967384 1 0.093874925 0.759307605
7 6 2 0.7351686 1 0.275875443 0.599417430
8 6 2 0.2139256 1 0.124149137 0.724577269
9 6 2 0.1161703 1 0.073665362 0.786072915
10 6 2 1.0400756 1 6.832203465 0.008952875
# Pearson's correlation test between each gene's expression values and the age of subjects
library(matrixTests)
row_cor_pearson(X, X$age)
obs.paired cor df statistic pvalue conf.low conf.high alternative cor.null conf.level
1 6 -0.5602433 4 -1.35270865 0.24755755 -0.9430338 0.46085636 two.sided 0 0.95
2 6 0.0384372 4 0.07693125 0.94237259 -0.7980176 0.82428560 two.sided 0 0.95
3 6 -0.9092607 4 -4.36906111 0.01197687 -0.9901613 -0.37278589 two.sided 0 0.95
4 6 -0.6950676 4 -1.93357802 0.12529880 -0.9632623 0.26724253 two.sided 0 0.95
5 6 -0.4096758 4 -0.89818427 0.41986507 -0.9165164 0.60205486 two.sided 0 0.95
6 6 -0.5186591 4 -1.21326433 0.29177292 -0.9361660 0.50580928 two.sided 0 0.95
7 6 -0.6143204 4 -1.55710099 0.19443841 -0.9515021 0.39334780 two.sided 0 0.95
8 6 -0.7763522 4 -2.46344001 0.06943424 -0.9741458 0.09517432 two.sided 0 0.95
9 6 0.7663925 4 2.38613079 0.07548443 -0.1194861 0.97285981 two.sided 0 0.95
10 6 -0.4486700 4 -1.00407581 0.37215468 -0.9238398 0.57069431 two.sided 0 0.95When needed 'annmatrix' can be transformed into a long-format data frame.
stack(X)
value chr pos group sex age
1:1 0.343167174 chr1 4186380 case F 10
2:1 1.101621650 chr1 4239287 case F 10
3:1 1.743441691 chr1 2480004 case F 10
4:1 1.474707248 chr1 2208404 case F 10
5:1 -0.083140482 chr2 4828541 case F 10
6:1 1.332632768 chr2 6622140 case F 10
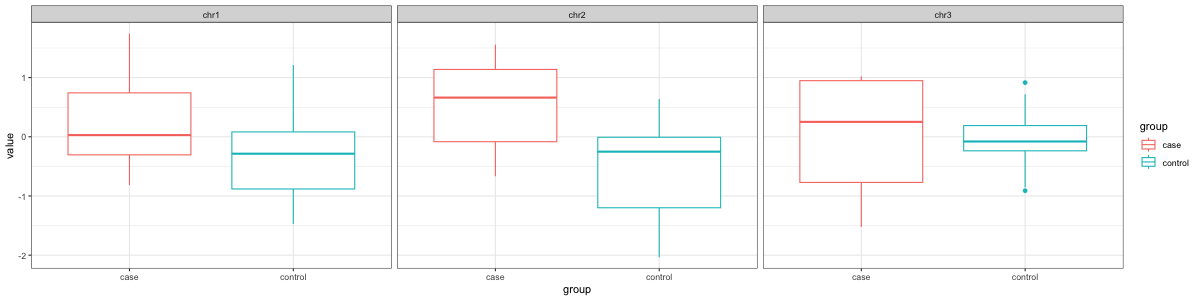
... ............ .... ....... .... . ..Which then can be used for things like making ggplot2 figures.
# case vs control boxplot by chromosome
library(ggplot2)
ggplot(stack(X), aes(x=group, y=value, color=group)) +
facet_wrap(~chr) +
geom_boxplot() +
theme_bw()annmatrix uses R's S3 class system to extend the base matrix class in order to provide it with persistent annotations that are associated with rows and columns.
Technically annmatrix object is just a regular R matrix with additional data.frame attributes .annmatrix.rann and .annmatrix.cann that are preserved after sub-setting and other matrix-specific operations.
As a result, every function that works on a matrix by design should work the same way with annmatrix.
Directly from CRAN:
install.packages("annmatrix")Or from GitHub, using remotes library:
remotes::install_github("karoliskoncevicius/annmatrix") # CRAN version
remotes::install_github("karoliskoncevicius/annmatrix@dev") # stable changes not yet on CRANSimilar ideas can be found in: