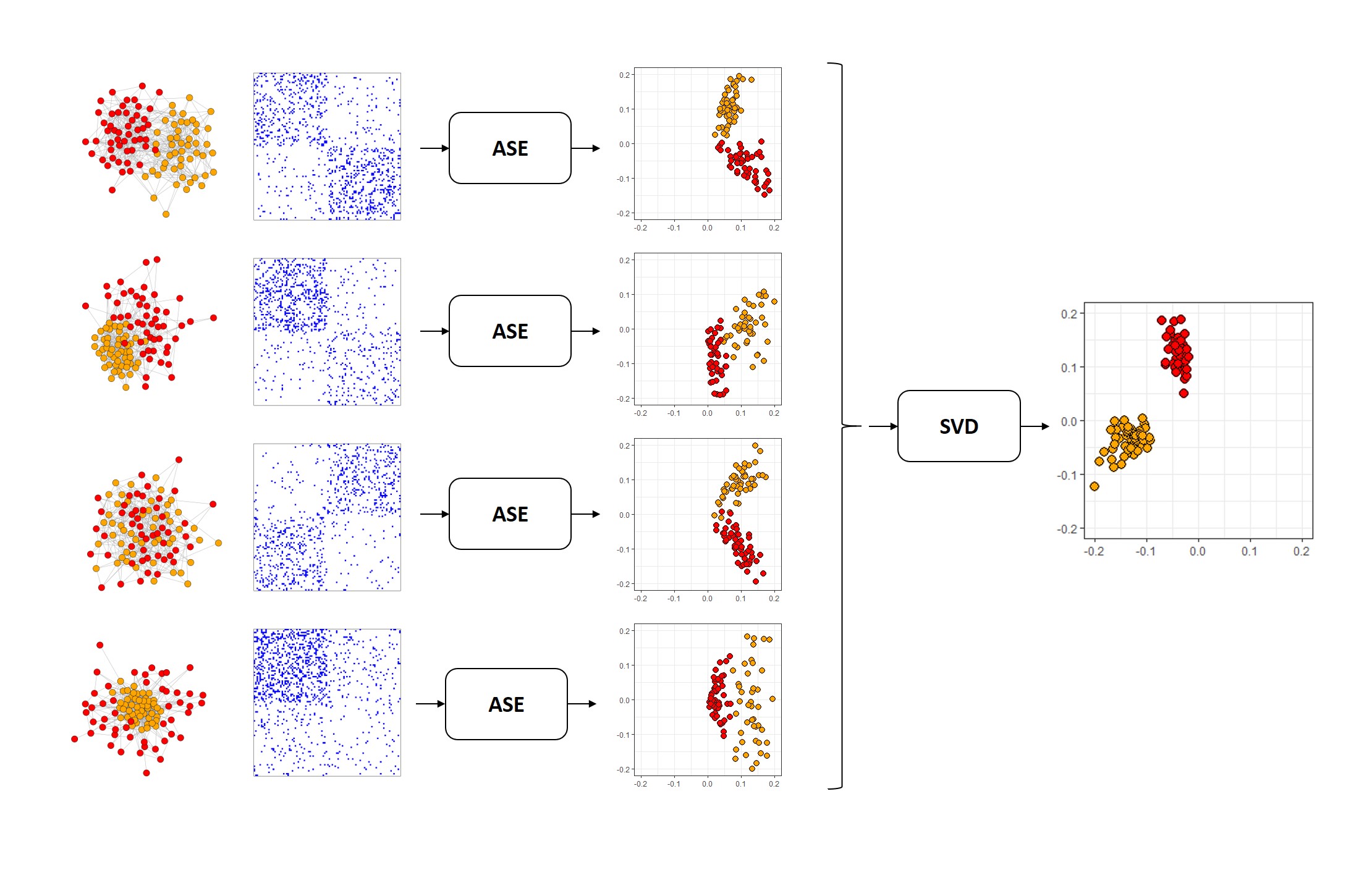
The multiple adjacency spectral embedding (MASE) performs a joint embedding of graphs based on a common invariant subspace assumption (see Arroyo et al. (2019)).
Given a sample of m graphs , each with n aligned vertices, MASE obtains a joint embedding of their adjacency matrices. The embedding of the graphs is based on the common subspace independent-edge (COSIE) random graph model, which assumes that the expected adjacency of all the graphs share the same invariant subspace of dimension
. Under this model, the expected adjacency of a graph
is given by
where
is a matrix with orthogonal columns that represents the basis of the common invariant subspace of the model, and
are individual score matrices that represent each graph.
To estimate the parameters of the embedding, MASE calculates a separate adjacency spectral embedding (ASE) for each graph, which consists in computing the eigendecomposition of each adjacency matrix, with a possible eigenvalue scaling, followed by a joint singular value decomposition of the concatenated ASEs. This procedure obtains an estimated common invariant subspace matrix (depicted in the picture below). After that, each adjacency matrix
is projected back to the eigenspace to obtain an estimated score matrix
The COSIE model and the MASE algorithm can be deployed for a number of subsequent network inference tasks, including
- Graph hypothesis testing
- Multiple graph dimensionality reduction
- Joint community detection on multiple networks
- Graph classification
This repository contains R code that implements MASE and other auxiliary functions. To use MASE, download all the content from the R/ folder. The file MASE-hyptest-example.R contains an example of MASE for two-sample graph hypothesis testing.
Arroyo, J., Athreya, A., Cape, J., Chen, G., Priebe, C.E., Vogelstein, J.T., "Inference for multiple heterogeneous networks with a common invariant subspace",