-
Notifications
You must be signed in to change notification settings - Fork 2
Home
This site provides semi-automated documentation of annotation extension relations and their usage, as well as a system for raising tickets to request improvements to these relations and their documentation. It includes:
- A tracker
- Semi-automated documentation of annotation extension relations
-
Automated reports of annotation extension usage
- These reports are generated by scripted SQL queries of the GOA database.
(Please note that documentation here has a technical focus and may not be complete. Original documentation still lives on the GOC wiki)
Many groups carrying out GO annotation restrict the meaning of GO terms used in annotation using annotation extensions (for full details please see Huntley et al., 2014). These extend a GO term with a relation and an object class. So, for example, rather than just asserting that a gene product is involved in 'sodium ion export from cell' a curator can record which cell type this occurs in as: 'occurs in' (some) 'motor neuron'.
Object classes may refer to a wide range of different types including cells, chemicals, proteins, genes, cellular components. Many of the relations used are also used by other OBO ontologies, and most are used in the full version of the gene ontology go-plus.owl, although some are specific to annotation extension. Using the same relations as in the full version of GO is critical to the proper interpretation of annotation extensions. So, for example, a query for proteins involved in processes occuring in motor neurons can find both the above annotation and annotations to processes that the ontology records as occuring in motor neurons.
Most annotation extension relations are shared with the full version of GO, but all have some attached axioms that are specific for annotation extension relations. We maintain all GO specific relations (but see #46), and GO specific axioms on external relations in gorel-edit.owl. This OWL file imports relations from the OBO Relations ontology. All relations used in annotation extensions are direct sub relations (sub objectProperties) of a grouping relation 'annotation extension relation' (GOREL_0000001). A Jenkins-based build process uses uses standard OWL-API module extraction methods (via owltools) to generate a relevant slice of ro.owl [gorel.obo], which is then merged with gorel-edit.owl minus imports to produced (http://purl.obolibrary.org/obo/go/extensions/gorel.obo) and gorel.owl. These files still contain more relations from RO than are actually used in annotation extensions. Subsets are used to tag currently valid relations (see below).

The main tool used for anotation extensions (and for annotation more generally) is Protein2GO. This displays only valid relations, groups annotation extensions by use and provides crude checks of their usage. Annotations made used Protein2GO are stored in the GOA database.
QuickGO has a graph of annotation extension relations, with floatover boxes showing key details and links to external documentation. Graph display is dependent on direct subrelation (subproperty) links between relations and so needs to display some grouping relations that are not used in annotation.
Annotation extension are checked for consistency/validity via 2 methods:
- Jenkins GAF checks include consistency checks on OWL interpretations of annotation extensions. For example, this annotation fails consistency checks because of classification based on the OWL domain of the relation used in annotation extension (results_in_maturation_of), combined with a disjointness declaration:
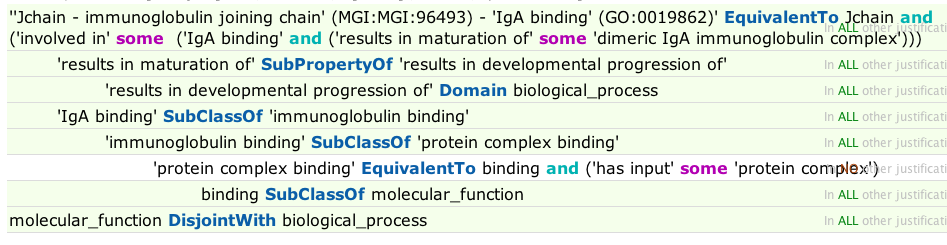
- QuickGO webservices check (mostly syntactic)
- What content is checked?
- Annotation extensions made in Protein2GO at the time of annotation, or when old annotations are reloaded.
- Old annotations failing checks are NOT loaded.
- Imported annoations with extensions in GAF/GPAD format.
- Failing extensions are NOT loaded into the DB. All such content is flushed and replaced nightly!
- All annotation extensions in the GOA database - as part of a monthly check.
- But in this case, a failure results only in a warning, content remains in the DB.
- Annotation extensions made in Protein2GO at the time of annotation, or when old annotations are reloaded.
- How is it checked?
- Is the relation in a known subset? (see below for details)
- Is usage consistent with local domain and range? (see below for details)
- What content is checked?
A set of subset tags are used by QuickGO webservices to assess validity for display, for use and to sort relations by usage. Loading checks are failed if at least one of these is not present (TBC):
-
display_for_curators: Display in the QuickGO graph.
-
extension relations begining with AE_ specificy crude grouping by range. The AE_ prefix is necessary as a way to distinguish subsets to be used for Annotation Extensions from those with other purposes that might come in from imports. A single relation may be in more than one grouping:
-
AE_biological_process
-
AE_cell_or_anatomical
-
AE_cellular_component
-
AE_chemical
-
AE_developmental_stages
-
AE_molecular_function
-
AE_sequence_feature
-
AE_sequence_or_complex
(Warning - the above list is up-to-date at the time of writing. Please check ontology files for the latest.)
(NOTE: this part of the infrastructure is under active development - see #13 for discussion, so this doc may go stale).
local_domain and local_range are annotation properties that allow a closed-world specification of the type of subject (domain) and object (range) allowed in annotation extension relations: If a subject or object class is not known to be a subclass of one of the classes listed in domain and range, then checks will fail. The value of local_domain must be a string consisting of a single OBO ID. The value of local_domain must a string consisting of a space-separated list of OBO identifiers.
Interpretation of domain uses the pre-reasoned GO graph and so is semantically robust. Interpretation of range is flakier - including, in some cases at least, a synatactic (ID string matching) component. Where the range covers only ontology terms, this is not necessary, as graph reasoning can be used. But where the range covers types that are not from ontologies (e.g. proteins, genes) it is necessary to attempt to check validity by checking if the ID used follows a known ID pattern for something of the specified type (e.g. a uniprot ID indicated a protein). This is not 100% reliable as some types of ID are ambiguous (examples TBA).
How is this achieved:
- A (pseudo) upper ontology, go-upper.obo, is used for basic classification.
- db-xrefs.yaml has mappings from external ID patterns to types in GO and go-upper. These classifications (via the graph) are used to determine validity of local domain and range.
- Some extra classification under BFO IDs is provided by a mapping table used by the QuickGO code. Ideally this will be replaced by a dynamically provided export of GO including classification under BFO, or failing that, classification under BFO in GO-upper.obo
DETAILS TO BE CHECKED & REFINED.