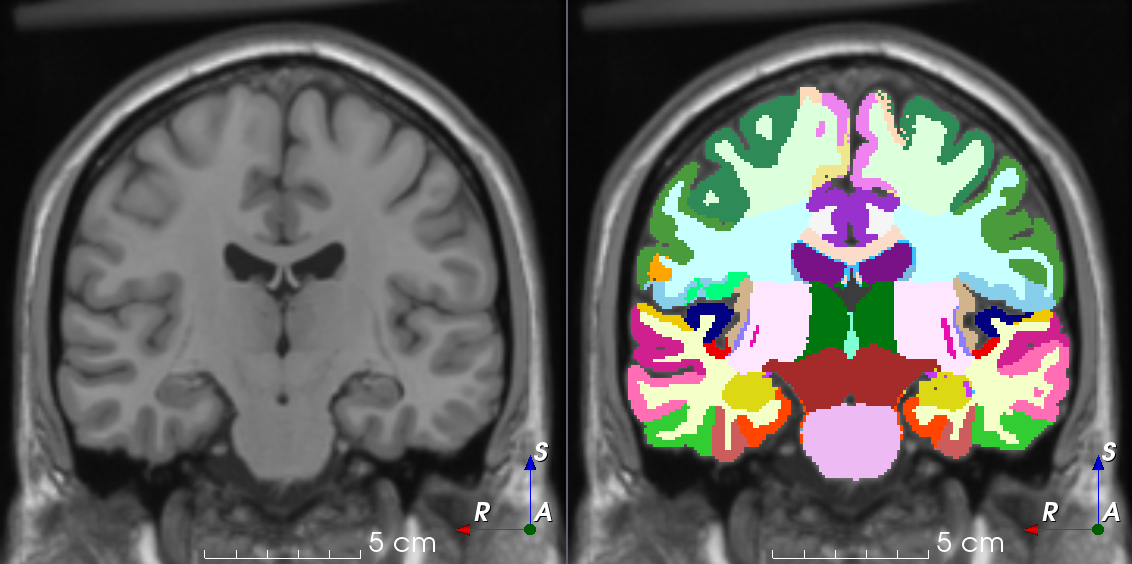
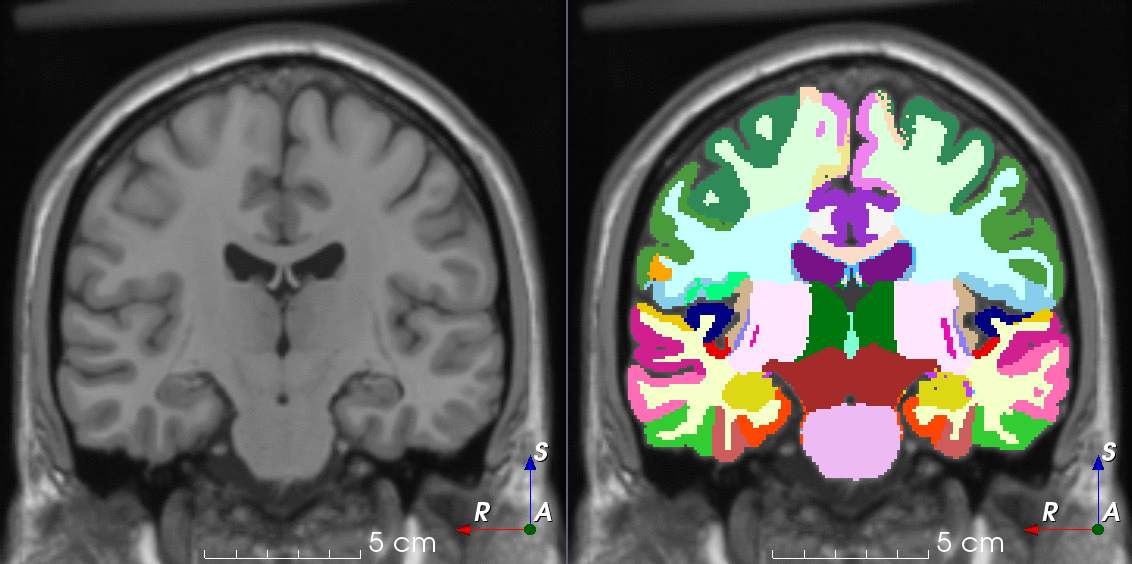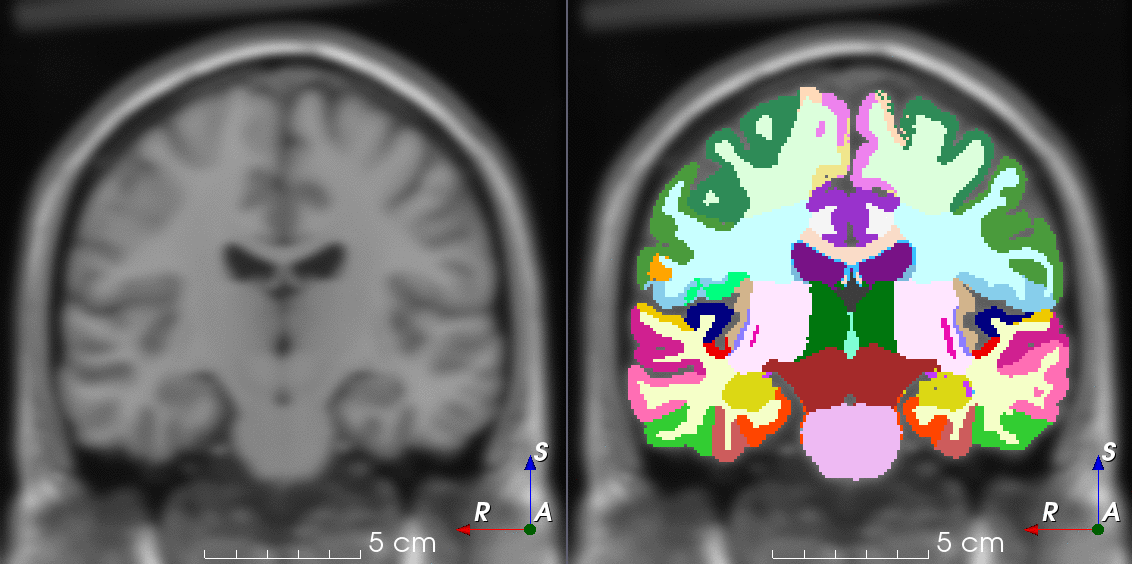
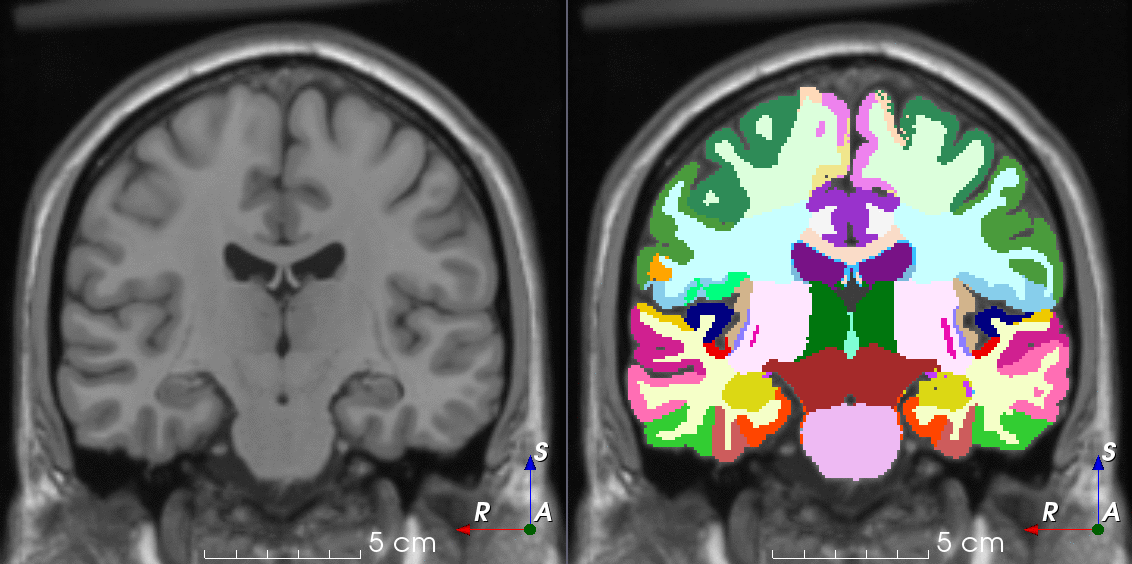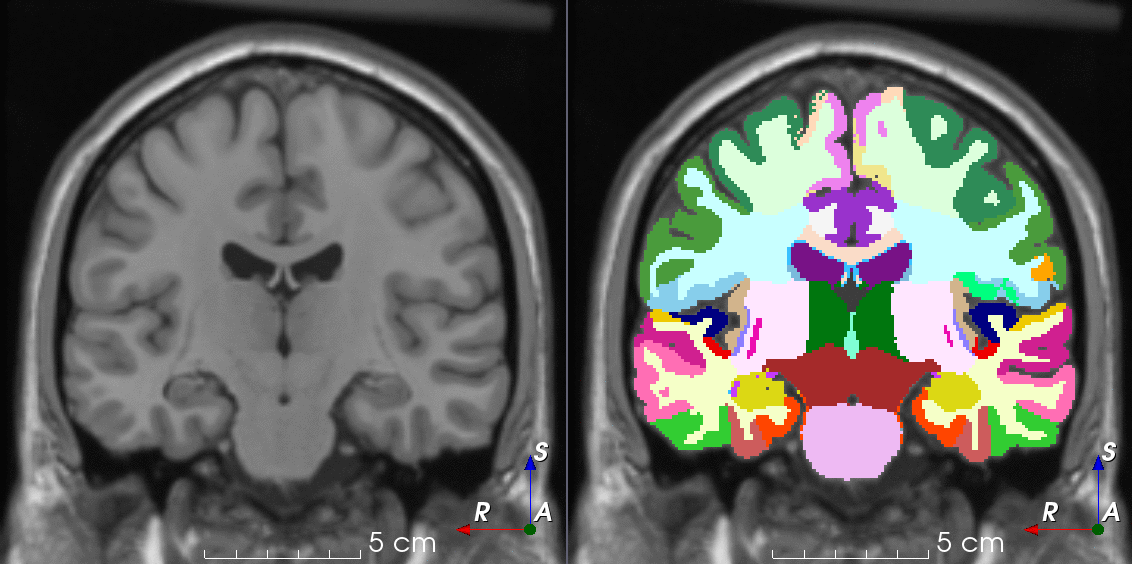
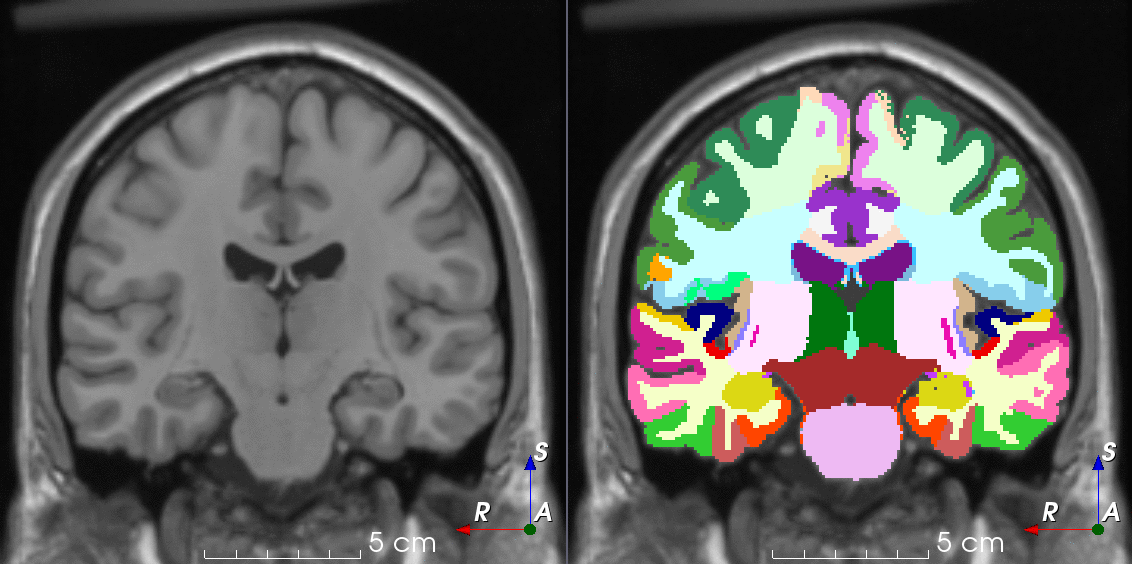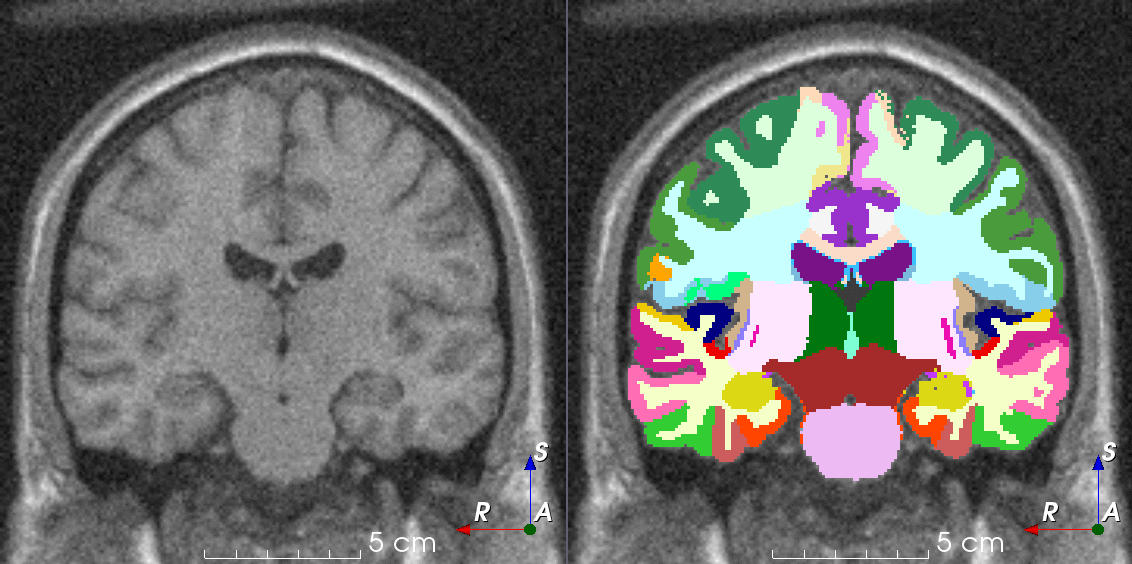
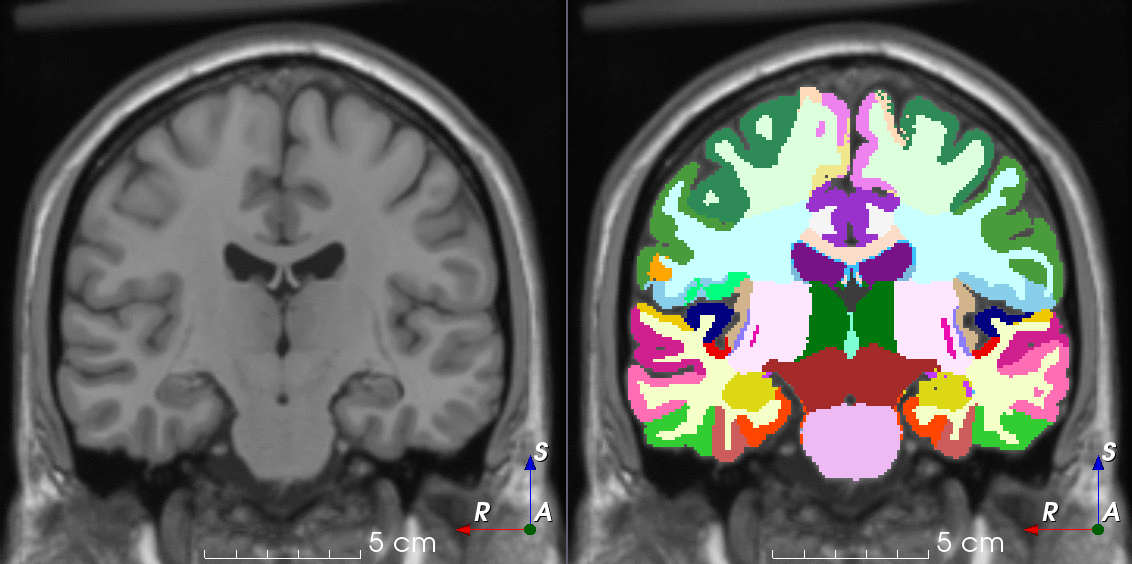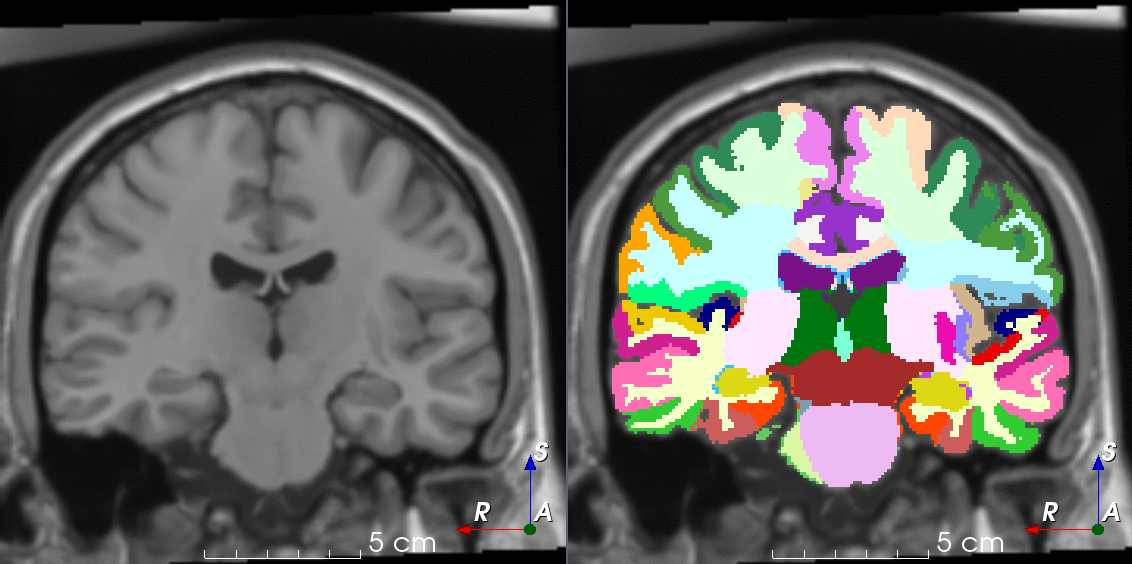
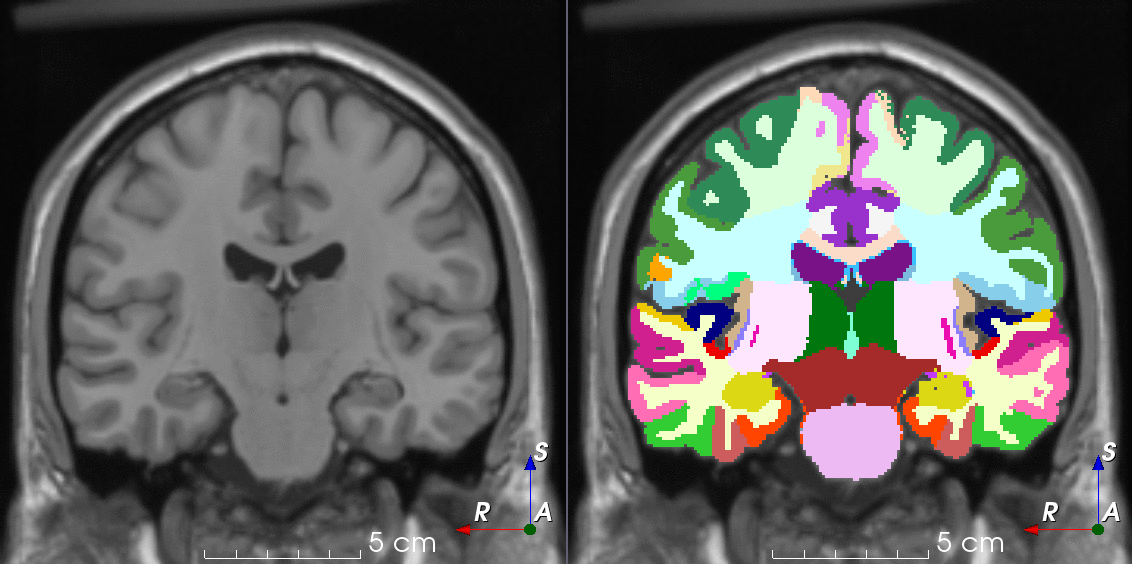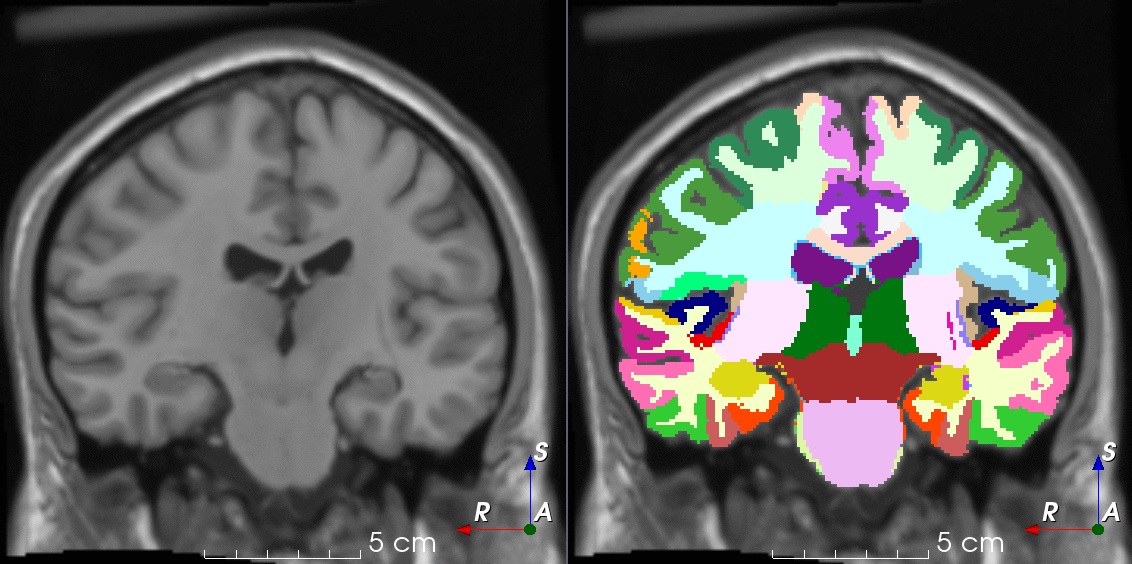
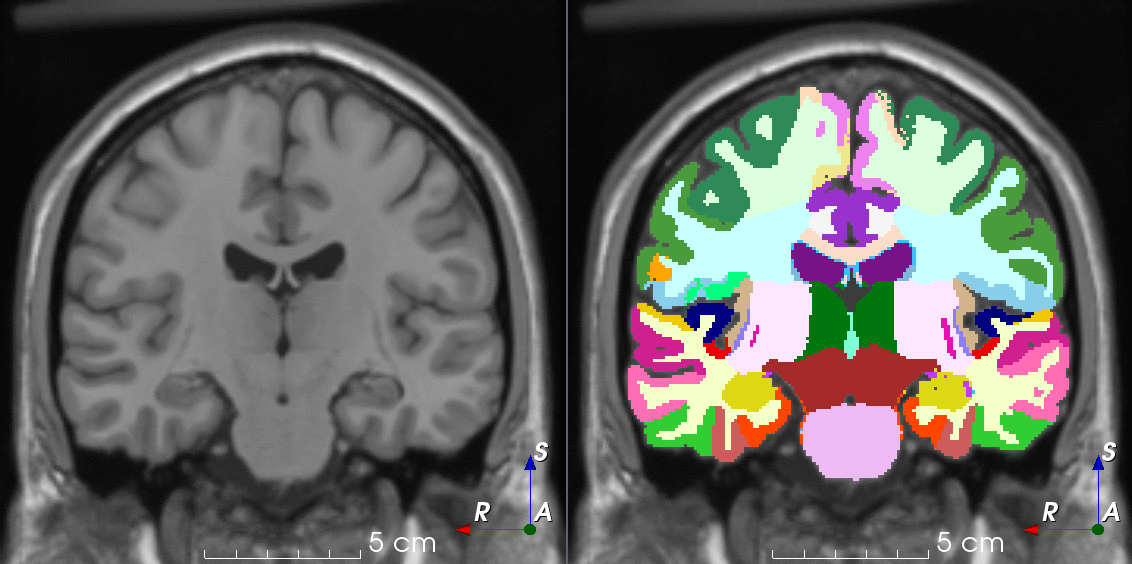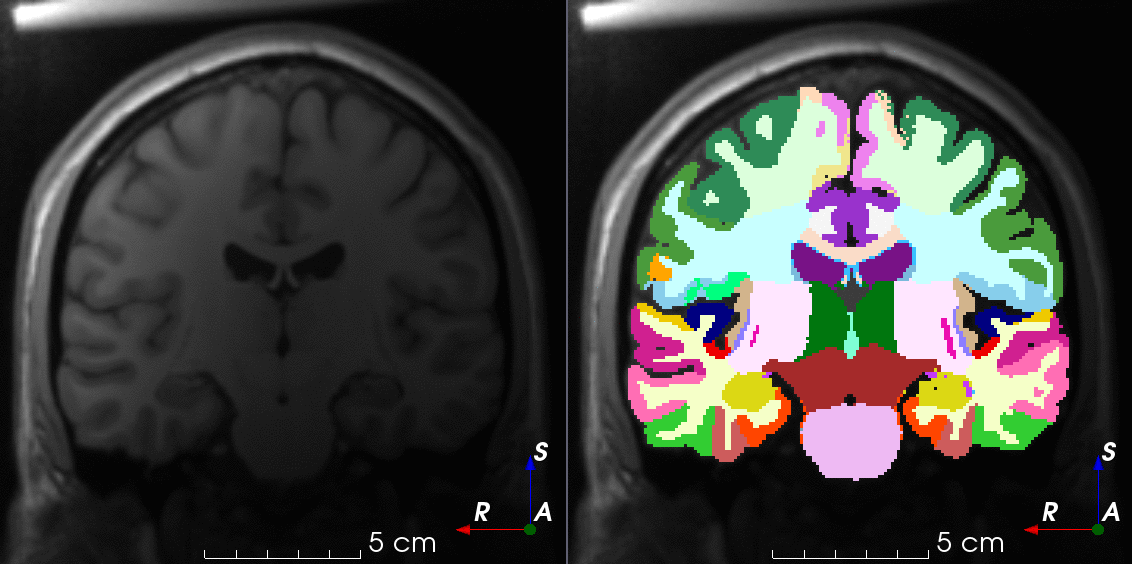
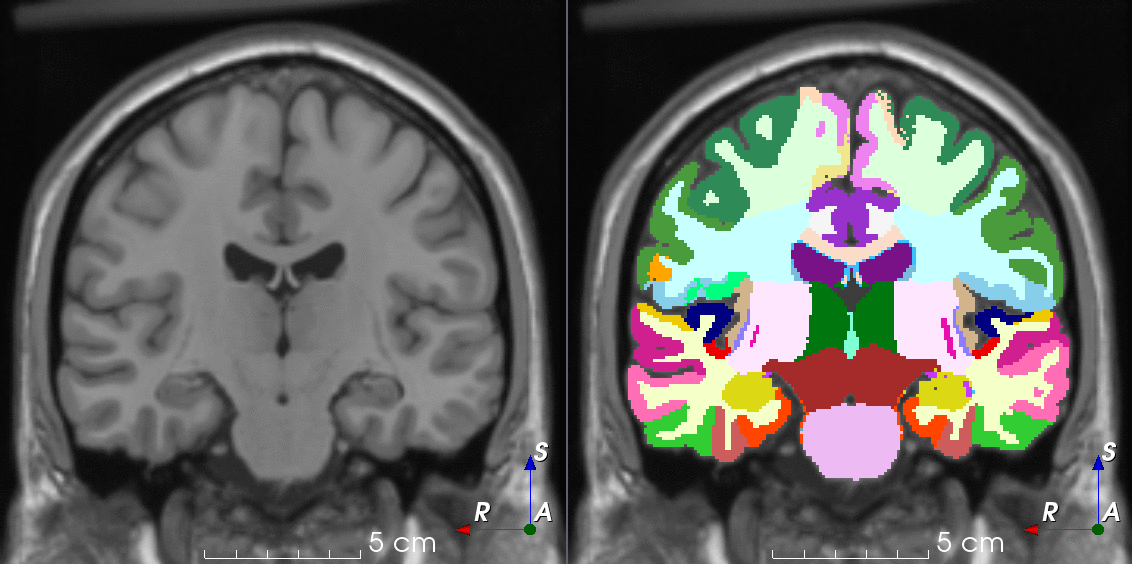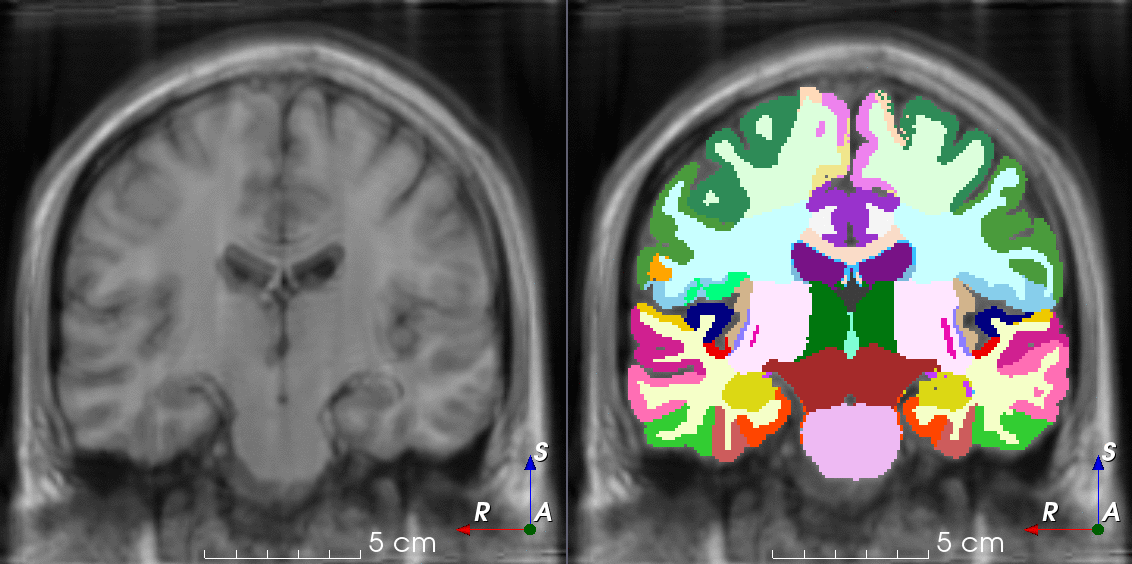
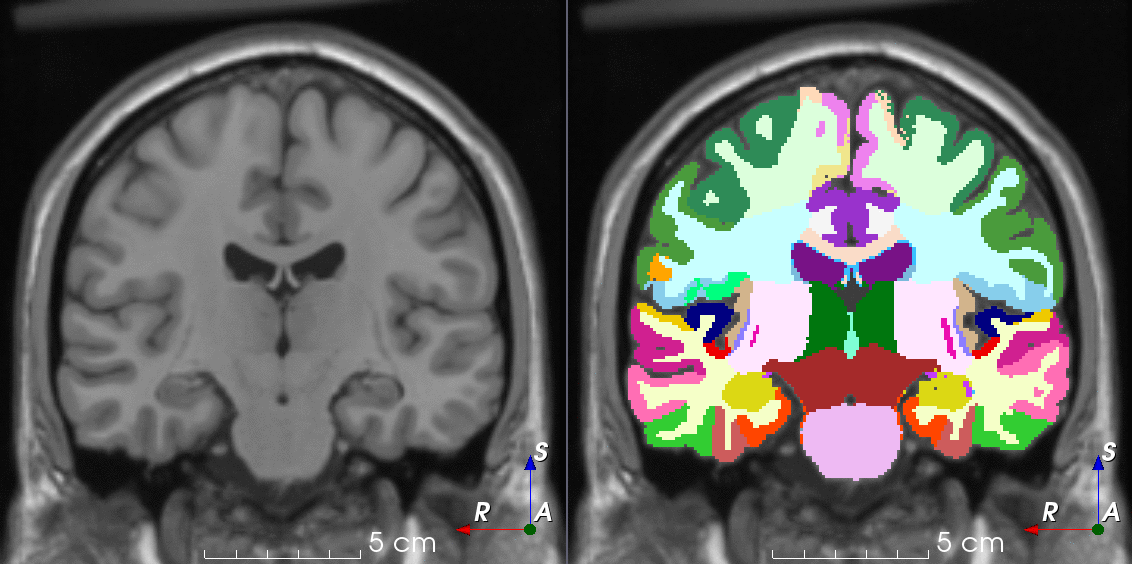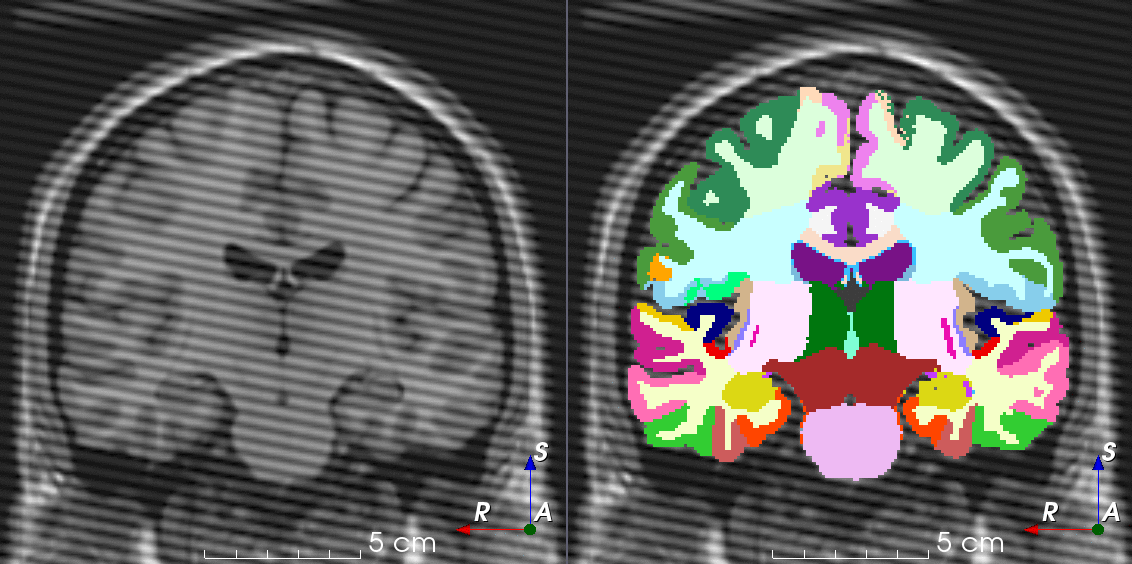
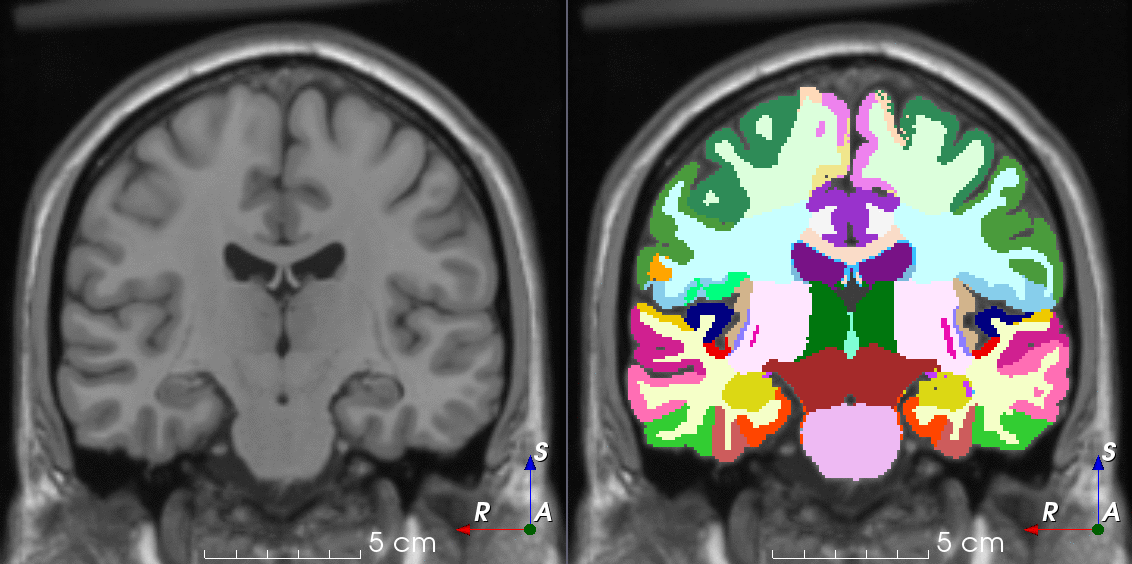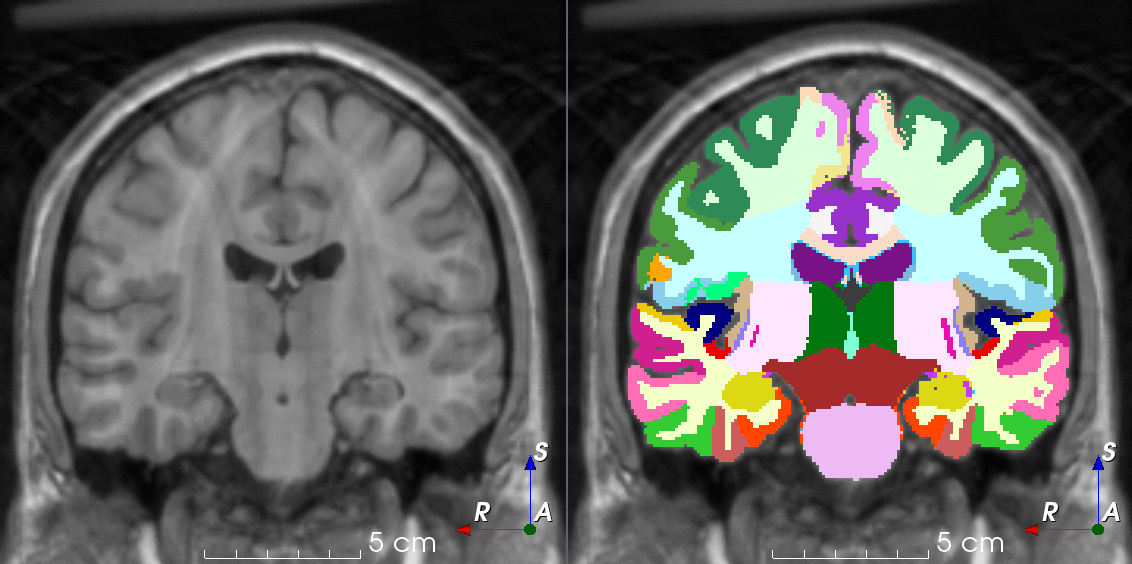
Tools like TorchIO are a symptom of the maturation of medical AI research using deep learning techniques.
Jack Clark, Policy Director at OpenAI (link).
This repository contains the code for a 3D Slicer extension that can be used to experiment with the TorchIO Python package without any coding.
More information on the extension can be found on the TorchIO documentation.
Please use the Preview Release of 3D Slicer.
The extension can be installed using Extensions Manager.
The build status can be checked on CDash.
If you like this repository, please click on Star!
If you use this tool for your research, please cite our paper:
BibTeX entry:
@article{perez-garcia_torchio_2021,
title = {TorchIO: a Python library for efficient loading, preprocessing, augmentation and patch-based sampling of medical images in deep learning},
journal = {Computer Methods and Programs in Biomedicine},
pages = {106236},
year = {2021},
issn = {0169-2607},
doi = {https://doi.org/10.1016/j.cmpb.2021.106236},
url = {https://www.sciencedirect.com/science/article/pii/S0169260721003102},
author = {P{\'e}rez-Garc{\'i}a, Fernando and Sparks, Rachel and Ourselin, S{\'e}bastien},
}This project is supported by the following institutions:
- Engineering and Physical Sciences Research Council (EPSRC) & UK Research and Innovation (UKRI)
- EPSRC Centre for Doctoral Training in Intelligent, Integrated Imaging In Healthcare (i4health) (University College London)
- Wellcome / EPSRC Centre for Interventional and Surgical Sciences (WEISS), University College London (UCL) (University College London)
- School of Biomedical Engineering & Imaging Sciences (BMEIS), King's College London (King's College London)