This code implements a generalization of the algorithm described in the paper "Efficient Projection onto the Perfect Phylogeny Model".
Please cite this code as:
@inproceedings{projection-onto-PPM, title={Efficient Projection onto the Perfect Phylogeny Model}, author={Jia, Bei and Ray, Surjyendu and Safavi, Sam and Bento, Jose}, booktitle={Advances in Neural Information Processing Systems}, year={2018} }
- To compile the code just do
gcc project_onto_PPM.c
- To run the code just do
./a.out InputDataExample OutputData 0
or
./a.out InputDataExample OutputData 1
-
The input data is of the following form, see the example file for exact formating instructions.
- number of nodes
- number of samples
- matrix of mutation frequencies, Fhat (column major form)
- vector that scales the norm in the objective
- root node
- vector of degree of each node in the tree
- adjancency list (neighbors of each node in the tree)
- flag, 0 or 1, that indicates whether to output the inferred fraction of each muutant, M, or just the projection cost, C(U)
-
When calling the program, the third argument, 0 or 1, indicates whether the columns of M must sum to 1, or sum to something smaller than 1.
-
If you want to test the code on random inputs, and compare against the output produced by CVX-Matlab, you can run the script test_against_CVX_matlab.m in Matlab, after you have installed CVX http://cvxr.com/cvx/download/
-
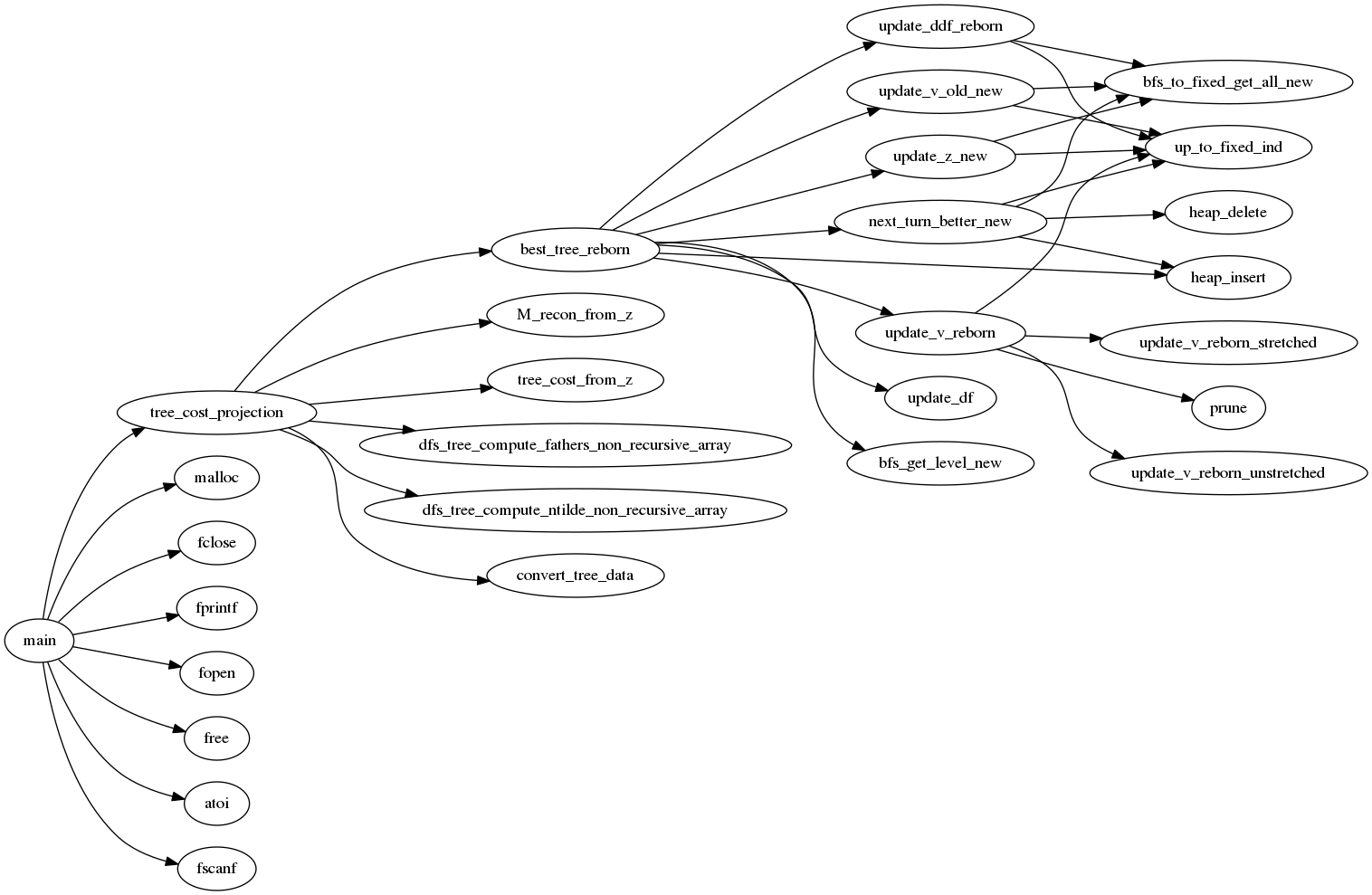
The dependencies of the different functions in the code is described by the following diagram.