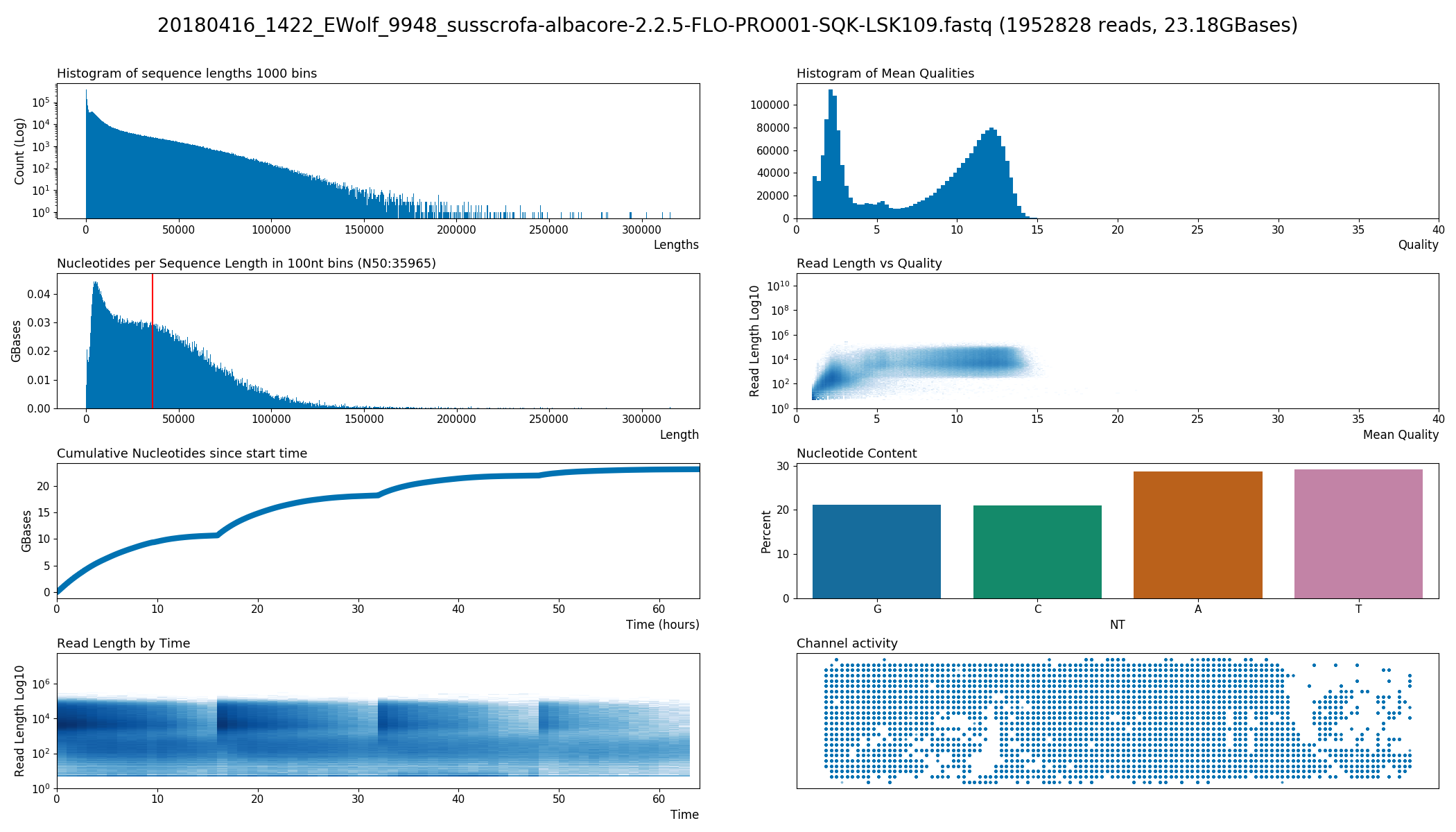
A collection of plots for long read sequencing FastQ files from devices like Oxford Nanopore's MinION and PromethION.
pip install --user --upgrade pandas seabornNote that matplot lib >= 3.0.0 is needed and 3.6.1 has a breaking bug (3.6.2 on are fine). Python >= 3.5 is therfore recommended, otherwise plots might be corrupted.
The aggregation of the data in python is quite slow. Therefore I implemented it as a standalone rust implementation, which can be found here: https://github.com/ahcm/lrdf
From PyPi:
pip3 install --user lrplotsFrom the release tar.gz:
python3 setup.py install --userWith the pip3 installation it should be in your PATH and you should be able to just call:
lrplot example.fastqIf the command can't be found, pip3 installed it in a path that is not in your PATH variable. You can then find it with:
find ~ -name lrplotor
find /usr/local -name lrplotThen either add the containing directory to your PATH or just use the full path.
You can also call it like a script with python3 (or whatever your python3 is called):
python3 bin/lrplots example.fastq