-
Notifications
You must be signed in to change notification settings - Fork 12
Commit
This commit does not belong to any branch on this repository, and may belong to a fork outside of the repository.
Merge pull request #108 from NREL/feature/calibration-demo
Feature/calibration demo
- Loading branch information
Showing
18 changed files
with
8,206 additions
and
126 deletions.
There are no files selected for viewing
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,59 @@ | ||
| # Calibration and Validation of Vehicle Models | ||
| FASTSim powertrain models can have varying levels of calibration and resolution based on available calibration and validation data. In the simplest (US) cases, the only available validation data for a powertrain model is the EPA "window sticker" energy consumption rates. However, there are also situations in which detailed dynamometer or on-road data is available for a particular vehicle, enabling much more detailed model calibration. This documentation is meant to summarize these various calibration levels and the tools available to help with more detailed calibration. | ||
|
|
||
| ## Calibration/Validation Levels | ||
|
|
||
| | Level | Calibration | Validation | | ||
| | --- | --- | --- | | ||
| | 0 | Vehicle is parameterized without any fitting to performance data. This is called __parameterization__, not calibration. | Could be none or could be validated against aggregate energy consumption data like EPA window sticker values. | | ||
| | 1 | Vehicle parameters are adjusted so that model results reasonably match test data for aggregate, cycle-level data (e.g. fuel usage, net SOC change). | Model results reasonably match at least some aggregate, cycle-level test data not used in any calibration process. | | ||
| | 2 | Vehicle parameters are adjusted so that model results reasonably match test data for time-resolved test data (e.g. instantaneous fuel usage, instantaneous cumulative fuel usage, instantaneous SOC). | Model results reasonably match at least some time-resolved test data not used in any calibration process. | | ||
| | 3 | Some amount of component-level thermal modeling is included and vehicle parameters are adjusted so that model results reasonably match test data for time-resolved test data (e.g. instantaneous fuel usage, instantaneous cumulative fuel usage, instantaneous SOC). | Model results reasonably match time-resolved test data not used in any calibration process that covers various temperatures and/vehcile transient thermal states. | | ||
|
|
||
| Examples of calibration levels 0, 2, and 3 from the [FASTSim Validation Report](https://www.nrel.gov/docs/fy22osti/81097.pdf): | ||
|
|
||
| 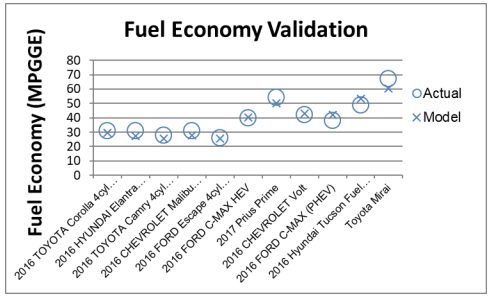 | ||
|
|
||
|  | ||
|
|
||
| 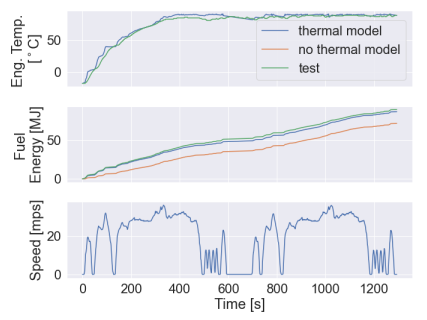 | ||
|
|
||
| ## Calibration Level 2 Guidelines | ||
| - Copy | ||
| [calibration_demo.py](https://github.com/NREL/fastsim/blob/fastsim-2/python/fastsim/demos/calibration_demo.py) | ||
| to your project directory and modify as needed. | ||
| - By default, this script selects the model that minimizes the euclidean error across | ||
| all objectives, which may not be the way that you want to select your final design. | ||
| By looking at the plots that get generated in `save_path`, you can use both the time | ||
| series and parallel coordinates plots to down select an appropriate design. | ||
| - Because PyMOO is a multi-objective optimizer that finds a multi-dimensional Pareto | ||
| surface, it will not necessarily return a single _best_ result -- rather, it will | ||
| produce a pareto-optimal set of results, and you must down select. Often, the design | ||
| with minimal euclidean error will be the best design, but it's good to pick a handful | ||
| of designs from the pareto set and check how they behave in the time-resolved plots | ||
| that can be optionally generated by the optimization script. | ||
| - Run `python calibration_demo.py --help` to see details about how to run calibration | ||
| and validation. Greater population size typically results in faster convergence at the | ||
| expense of increased run time for each generation. There's no benefit in having a | ||
| number of processes larger than the population size. `xtol` and `ftol` (see CLI help) | ||
| can be used to adjust when the minimization is considered converged. If the | ||
| optimization is terminating when `n_max_gen` is hit, then that means it has not | ||
| converged, and you may want to increase `n_max_gen`. | ||
| - Usually, start out with an existing vehicle model that is reasonably close to the | ||
| new vehicle, and make sure to provide as many explicit parameters as possible. In | ||
| some cases, a reasonable engineering judgment is appropriate. | ||
| - Resample data to 1 Hz. This is a good idea because higher frequency data will cause | ||
| fastsim to run more slowly. This can be done with `fastsim.resample.resample`. Be | ||
| sure to specify `rate_vars` (e.g. fuel power flow rate [W]), which will be time | ||
| averaged over the previous time step in the new frequency. | ||
| - Identify test data signals and corresponding fastsim signals that need to match. | ||
| These pairs of signals will be used to construct minimization objectives. See | ||
| where `obj_names` is defined in `calibration_demo.py` for an example. | ||
| - See where `cycs[key]` gets assigned to see an example of constructing a Cycle from a dataframe. | ||
| - Partition out calibration/validation data by specifying a tuple of regex patterns | ||
| that correspond to cycle names. See where `cal_cyc_patterns` is defined for an | ||
| example. Typically, it's good to reserve about 25-33% of your data for validation. | ||
| - To set parameters and corresponding ranges that the optimizer is allowed to use in | ||
| getting the model to match test data, see where `params_and_bounds` is defined | ||
| below. | ||
|
|
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
Oops, something went wrong.