-
Notifications
You must be signed in to change notification settings - Fork 85
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Lsim gpu #468
Conversation
|
One other thing is that if you are using GPU you currently have to send |
|
This is an automated message.
|
| # Using similar instead of Matrix{T} to allow for CuArrays to be used. | ||
| # This approach is problematic if x0 is sparse for example, but was considered | ||
| # to be good enough for now | ||
| x = similar(x0, T, (length(x0), n)) | ||
|
|
||
| x[:,1] .= x0 | ||
| mul!(x[:, 2:end], B, transpose(u[1:end-1, :])) # Do all multiplications B*u[:,k] to save view allocations |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Was there a problem with mul! on the GPU? Otherwise the mul! with views should avoid the allocation.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
There was something with the views that made me remove it. Will have a look at what was going on.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
I think your benchmark might be biased since you input u as a transpose btw
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Worse is that the array is not strided when using views due to the removal of the last row.
Transposes are not always less efficient, A'B is usually more efficient than AB.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
I think that we've already had some offline discussions regarding the intput/output format of lsim, but I think that we decided to push the decision to the future. I opened a new issue instead of hijacking this thread, see #469.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
@mfalt Yeah, makes sense. Totally missed that.
Trying some different versions of this specific line.
First calling lsim with discrete sys and u vec where (ny, nx, nu) = (100, 500, 200) and u vec is not a transpose anymore.
@views mul!(x[:, 2:end], B, transpose(u[1:end-1, :]))error on GPU, 4.2ms median on CPU@views mul!(x[:, 2:end], B, transpose(u)[:, 1:end-1])3.2s medain on GPU, 13.7ms median on CPUx[:, 2:end] .= B * transpose(u[1:end-1, :])2.7ms median on GPU, 4.8ms median on CPUx[:, 2:end] .= B * transpose(u)[:, 1:end-1]287ms median on GPU, 4.8ms median on CPU
The calling lsim with (nx, ny, nu) = (1, 5, 2) (only for CPU here).
@views mul!(x[:, 2:end], B, transpose(u[1:end-1, :]))31micros median on CPU@views mul!(x[:, 2:end], B, transpose(u)[:, 1:end-1])35micros median on CPUx[:, 2:end] .= B * transpose(u[1:end-1, :])24micros median on CPUx[:, 2:end] .= B * transpose(u)[:, 1:end-1]24micros median on CPU
So the original with views and mul! does not work with GPU for some reason, I have not dug deeper into why. Of the others it seems like the GPU does not like to transpose a sliced matrix (100x slower) so that is why I switched it around. And this held even when not sending in the u vec as a transpose.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
I think the problem here is the data layout as discussed in #469
We need to chop u in a very unfortunate way, either to make a new copy and pay the allocation as in the last two examples, or use a view into a non-strided array, probably causing a generic fallback method to be called rather than an optimized BLAS/CUBLAS method. had the memory layout been transposed, i.e., u in R(nu x T), we could use @view and get a strided array hitting a BLAS method and both GPU and CPU versions would be blazing fast without allocations.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Example
x = zeros(4, 2^10)
u = zeros(4, 2^10)
B = randn(4,4)
@btime @views mul!($(x)[:, 2:end], $(B), $(u)[:, 2:end]) # 3.873 μs (0 allocations: 0 bytes)
@btime @views mul!($(x)[:, 2:end], $(B), $(u')[2:end, :]') # 29.313 μs (6 allocations: 336 bytes)There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Yeah, I don't know what the best solution would be.
If #469 will happen then I think that would solve the problem.
I also asked in the CUDA.jl community and was told that the array type was too complex for them to do any specialized dispatch (mul! of a view of a transpose of a slice) but was told that this PR JuliaGPU/GPUArrays.jl#352 would allow us to get it working but not optimized for GPU using the current CPU-optimized code.
|
Closing this, most of this was updated in #480 |
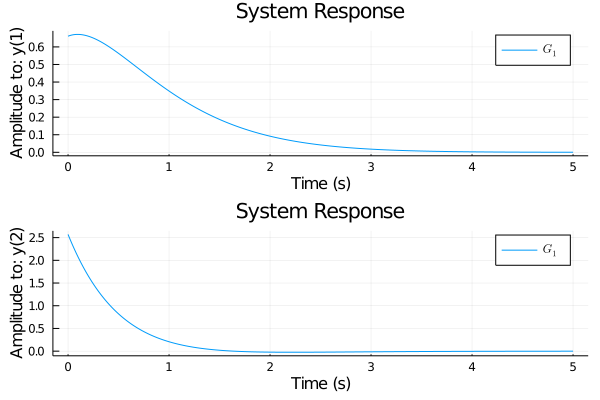
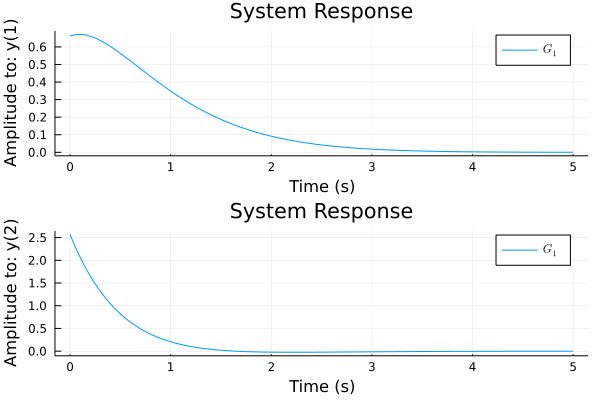
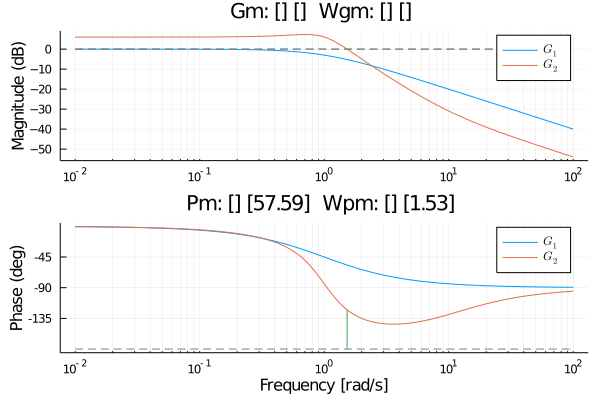
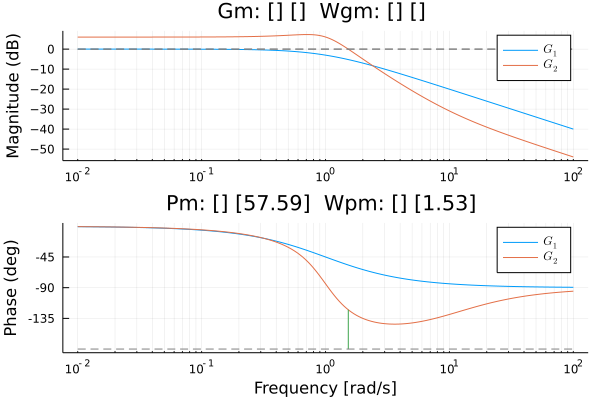
Changed some lsim and ltitr code so it will support CuArrays or HeteroStateSpace with CuArrays.
Ran this test on master and new branch to make sure it kept same performance on normal arrays.
Results on master
Results on new branch
This code was used to compare normal system and ones using CuArrays
which gave this result