This R Shiny application can be used to predict Gene Ontology (GO) terms indirectly targeted by human microRNAs. In contrast to direct targets which are predicted to have binding sites, indirect targets are regulated transcriptionally through transcription factors (TFs).
There are two ways to run this tool,
can be accessed directly from this link https://fadeel.shinyapps.io/miRinGO
- Download R shiny app source code from GitHub https://github.com/Fadeel/miRinGO/archive/master.zip
- Open
miRinGO.Rin RStudio. - Make sure the following packages are installed (shinythemes, shiny, DT, dplyr, stringr, ggplot2,tm and wordcloud)
- You can run the application by clicking the 'Run App' button
| Parameter | Description | Possible values |
|---|---|---|
| Tissue type | 29 different tissues | Brain, colon, lung, .. |
| input miRNAs | miRBase miRNA ID | hsa-miR-9-5p |
| GO category | Choose the category of GO annotation | Biological process OR cellular component OR molecular function |
| Targeting mode | Choose mode of miRNA targeting | Direct OR indirect |
| Percentage of TargetScan target genes | percentage of top-ranked targeted genes (sorted by TargetScan v7.2 context++ score) | [20% - 100%], step size 20% |
| min. number of genes per GO term | GO terms with number of genes less than this number will be removed | integer value (default is 5) |
| Number of GO terms/pathways for visualization | Number of GO terms/pathways to be used for bar plot and word cloud | integer value (default is 10) |
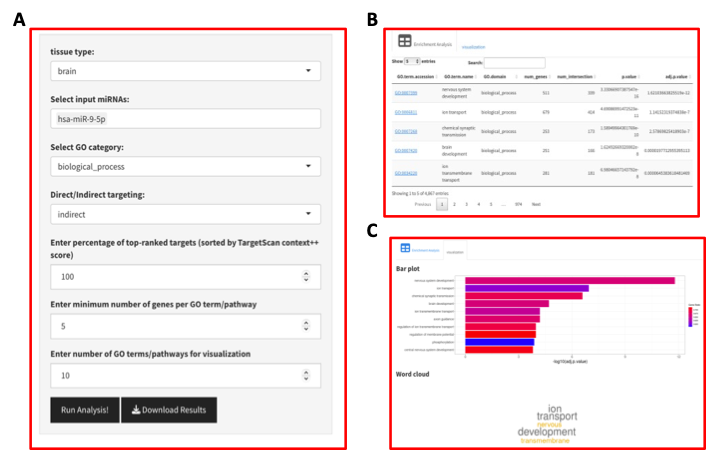
This app has two panels, left one for input data and parameters selection and the right one for displaying the results as shown in below (a) input parameters; (b) Table with miRNA GO enrichment analysis; (c) Bar plot and word cloud summarizing the top enriched GO terms.