-
Notifications
You must be signed in to change notification settings - Fork 749
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
2D time-dependent heat equation #61
Comments
|
See "Q: I failed to train the network, e.g., the training loss is large." at "Questions & Answers" at the webpage. Note: |
|
I think you should normalize you PDE or apply network output transform, the scales of your parameters differ a lot. |
|
@lululxvi @qizhang94 Thanks a lot for your answers. I have tried to scale the problem. Since the solution for this problem is between [300, 1000]. I have rescaled the problem by 100. In addition, I have also tried to use the loss_weight to rescale the weight. However, the problem does not converge. Do you have any suggestions? The updated code is provided: mport numpy as np
import tensorflow as tf
import matplotlib.pyplot as plt
import deepxde as dde
from scipy.interpolate import griddata
import matplotlib.gridspec as gridspec
from mpl_toolkits.mplot3d import axes3d
# geometry parameters
xdim = 200
ydim = 100
xmin = 0.0
ymin = 0.0
xmax = 0.04
ymax = 0.02
# input parameters
rho = 8000
cp = 500
k = 200
T0 = 300
Tinit = 1000
t0 = 0.0
te = 30.0
x_start = 0.0 # laser start position
# dnn parameters
num_hidden_layer = 3 # number of hidden layers for DNN
hidden_layer_size = 40 # size of each hidden layers
num_domain=1000 # number of training points within domain Tf: random points (spatio-temporal domain)
num_boundary=1000 # number of training boundary condition points on the geometry boundary: Tb
num_initial= 1000 # number of training initial condition points: Tb
num_test=None # number of testing points within domain: uniform generated
epochs=10000 # number of epochs for training
lr=0.001 # learning rate
def main():
def pde(x, T):
dT_x = tf.gradients(T, x)[0]
dT_x, dT_y, dT_t = dT_x[:,0:1], dT_x[:,1:2], dT_x[:,2:]
dT_xx = tf.gradients(dT_x, x)[0][:, 0:1]
dT_yy = tf.gradients(dT_y, x)[0][:, 1:2]
return rho*cp*dT_t - k*dT_xx - k*dT_yy
def boundary_x_l(x, on_boundary):
return on_boundary and np.isclose(x[0], xmin)
def boundary_x_r(x, on_boundary):
return on_boundary and np.isclose(x[0], xmax)
def boundary_y_b(x, on_boundary):
return on_boundary and np.isclose(x[1], ymin)
def boundary_y_u(x, on_boundary):
return on_boundary and np.isclose(x[1], ymax)
def func(x):
return np.ones((len(x),1), dtype=np.float32)*T0
def func_n(x):
return np.zeros((len(x),1), dtype=np.float32)
def func_init(x):
return np.ones((len(x),1), dtype=np.float32)*Tinit
geom = dde.geometry.Rectangle([0, 0], [xmax, ymax])
timedomain = dde.geometry.TimeDomain(t0, te)
geomtime = dde.geometry.GeometryXTime(geom, timedomain)
bc_x_l = dde.DirichletBC(geomtime, func, boundary_x_l)
bc_x_r = dde.DirichletBC(geomtime, func, boundary_x_r)
bc_y_b = dde.DirichletBC(geomtime, func, boundary_y_b)
bc_y_u = dde.NeumannBC(geomtime, func_n, boundary_y_u)
ic = dde.IC(geomtime, func_init, lambda _, on_initial: on_initial)
data = dde.data.TimePDE(
geomtime,
pde,
[bc_x_l, bc_x_r, bc_y_b, bc_y_u, ic],
num_domain=num_domain,
num_boundary=num_boundary,
num_initial=num_initial,
# train_distribution="uniform",
num_test=num_test
)
net = dde.maps.FNN([3] + [hidden_layer_size] * num_hidden_layer + [1], "tanh", "Glorot uniform")
# net.apply_output_transform(lambda x, y: y*1000)
net.apply_output_transform(lambda x, y: y*100)
model = dde.Model(data, net)
model.compile("adam", lr=lr, loss_weights=[1e-12, 1e-2, 1e-2, 1e-2, 1, 1e-3])
# model.compile("adam", lr=lr)
losshistory, train_state = model.train(epochs=epochs)
# model.compile("L-BFGS-B")
# losshistory, train_state = model.train()
dde.saveplot(losshistory, train_state, issave=False, isplot=False)
if __name__ == "__main__":
main() |
|
Try to train for more iterations. |
@lululxvi, I have tried your suggestion, but the problem still does not converge (see the attached picture): |
|
There are some problems of the scaling:
|
|
@lululxvi I Thanks for your quick response. have tried different orders, i.e. 100 and 1000 using the |
|
Hello @wangcj05 ! I am trying to solve a similar problem. Did you achieve convergence? |
|
@Huzaifg Unfortunately, I do not get the problem converged. |
|
@wangcj05 I actually got this to work a while back but forgot to update here. Given below is the complete code import deepxde as dde
import matplotlib.pyplot as plt
import numpy as np
from deepxde.backend import tf
# Some useful functions
t1 = 0
t2 = 1
end_time = 1
def pde(X,T):
dT_xx = dde.grad.hessian(T, X ,j=0)
dT_yy = dde.grad.hessian(T, X, j=1)
dT_t = dde.grad.jacobian(T, X, j=2)
# Dividing by rhoc to make it 1
rhoc = (3.8151 * 10**3) / (3.8151 * 10**3)
kap = (385 / (3.8151 * 10**3))
# no forcing function
return ((rhoc * dT_t) - (kap * (dT_xx + dT_yy)))
def r_boundary(X,on_boundary):
x,y,t = X
return on_boundary and np.isclose(x,1)
def l_boundary(X,on_boundary):
x,y,t = X
return on_boundary and np.isclose(x,0)
def up_boundary(X,on_boundary):
x,y,t = X
return on_boundary and np.isclose(y,1)
def down_boundary(X,on_boundary):
x,y,t = X
return on_boundary and np.isclose(y,0)
def boundary_initial(X, on_initial):
x,y,t = X
return on_initial and np.isclose(t, 0)
def init_func(X):
x = X[:, 0:1]
y = X[:, 1:2]
t = np.zeros((len(X),1))
for count,x_ in enumerate(x):
if x_ < 0.5:
t[count] = t1
else:
t[count] = t1 + (2) * (x_ - 0.5)
return t
def dir_func_l(X):
return t1 * np.ones((len(X),1))
def dir_func_r(X):
return t2 * np.ones((len(X),1))
def func_zero(X):
return np.zeros((len(X),1))
def hard(X, T):
x,y,t = x[:, 0:1], x[:, 1:2],x[:,2:3]
return (r - r_in) * y + T_star
num_domain = 30000
num_boundary = 8000
num_initial = 20000
layer_size = [3] + [60] * 5 + [1]
activation_func = "tanh"
initializer = "Glorot uniform"
lr = 1e-3
# Applying Loss weights as given below
# [PDE Loss, BC1 loss - Dirichlet Left , BC2 loss - Dirichlet Right, BC3 loss- Neumann up, BC4 loss - Neumann down, IC Loss]
loss_weights = [10, 1, 1, 1, 1, 10]
epochs = 10000
optimizer = "adam"
batch_size_ = 256
geom = dde.geometry.Rectangle(xmin=[0, 0], xmax=[1, 1])
timedomain = dde.geometry.TimeDomain(0, end_time)
geomtime = dde.geometry.GeometryXTime(geom, timedomain)
bc_l = dde.DirichletBC(geomtime, dir_func_l, l_boundary)
bc_r = dde.DirichletBC(geomtime, dir_func_r, r_boundary)
bc_up = dde.NeumannBC(geomtime, func_zero, up_boundary)
bc_low = dde.NeumannBC(geomtime, func_zero, down_boundary)
ic = dde.IC(geomtime, init_func, boundary_initial)
data = dde.data.TimePDE(
geomtime, pde, [bc_l, bc_r, bc_up, bc_low, ic], num_domain=num_domain, num_boundary=num_boundary, num_initial=num_initial)
net = dde.maps.FNN(layer_size, activation_func, initializer)
net.apply_output_transform(lambda x, y: abs(y))
## Uncomment below line to apply hard Dirichlet Boundary Conditions
# net.outputs_modify(lambda x, y: x[:,0:1]*t2 + x[:,0:1] * (1 - x[:,0:1]) * y)
model = dde.Model(data, net)
model.compile(optimizer, lr=lr,loss_weights=loss_weights)
# To save the best model every 1000 epochs
checker = dde.callbacks.ModelCheckpoint(
"model/model1.ckpt", save_better_only=True, period=1000
)
losshistory, trainstate = model.train(epochs=epochs,batch_size = batch_size_,callbacks = [checker])
model.compile("L-BFGS-B")
dde.optimizers.set_LBFGS_options(
maxcor=50,
)
losshistory, train_state = model.train(epochs = epochs, batch_size = batch_size_)
dde.saveplot(losshistory, trainstate, issave=True, isplot=True)The best results achieved, import matplotlib.animation as animation
from matplotlib.animation import FuncAnimation
ax = fig.add_subplot(111)
nelx = 100
nely = 100
timesteps = 101
x = np.linspace(0,1,nelx+1)
y = np.linspace(0,1,nely+1)
t = np.linspace(0,1,timesteps)
delta_t = t[1] - t[0]
xx,yy = np.meshgrid(x,y)
x_ = np.zeros(shape = ((nelx+1) * (nely+1),))
y_ = np.zeros(shape = ((nelx+1) * (nely+1),))
for c1,ycor in enumerate(y):
for c2,xcor in enumerate(x):
x_[c1*(nelx+1) + c2] = xcor
y_[c1*(nelx+1) + c2] = ycor
Ts = []
for time in t:
t_ = np.ones((nelx+1) * (nely+1),) * (time)
X = np.column_stack((x_,y_))
X = np.column_stack((X,t_))
T = model.predict(X)
T = T*30
T = T.reshape(T.shape[0],)
T = T.reshape(nelx+1,nely+1)
Ts.append(T)
def plotheatmap(T,time):
# Clear the current plot figure
plt.clf()
plt.title(f"Temperature at t = {time*delta_t} unit time")
plt.xlabel("x")
plt.ylabel("y")
plt.pcolor(xx, yy, T,cmap = 'RdBu_r')
plt.colorbar()
return plt
def animate(k):
plotheatmap(Ts[k], k)
anim = animation.FuncAnimation(plt.figure(), animate, interval=1, frames=len(t), repeat=False)
anim.save("trial1.gif")You can also find the gif converted to a mp4 file at the Gdrive link Please do let me know if I could have done something better or some mistakes that you find, I am only starting out and would love to learn! |
|
@Huzaifg Thank you for an excellent explanation and code upload. Works perfectly. thanks again. |
|
Hey @MINE0126 Can you post your loss vs Epochs graph for the above result? |
|
Hi~ @MINE0126 Would you mind posting the correct code here? Thans in advance! |
|
@Huzaifg Hi. `a1=1.67e-4 geom = dde.geometry.Rectangle(xmin=[0,0], xmax=[5,5]) Thanks! |
|
Hello @123new-net , |
|
Hi~ @Huzaifg The initial function needs to be 2 dimensional. But you just use "np.zeros((len(X),1))" here. So I'm wondering about this. Hope for your reply. Thanks! |
|
Hey @Huzaifg, Your code above has been really helpful. Thank you for sharing with the community. I am quite new to DeepXDE, and have a few doubts. I was wondering if you might have any answers for them.
Hope for your reply. |
Hi |
|
Hello everyone, Does anyone have any experience with plotting the above solution Thank you in advance. |
here if we reduce num_initial points to 400. then we get absurd result. so can you tell the approach to solve 2d heat equation with less initial points |







Hello,
I tried to using DeepXDE to solve 2D time-dependent heat equations, i.e.
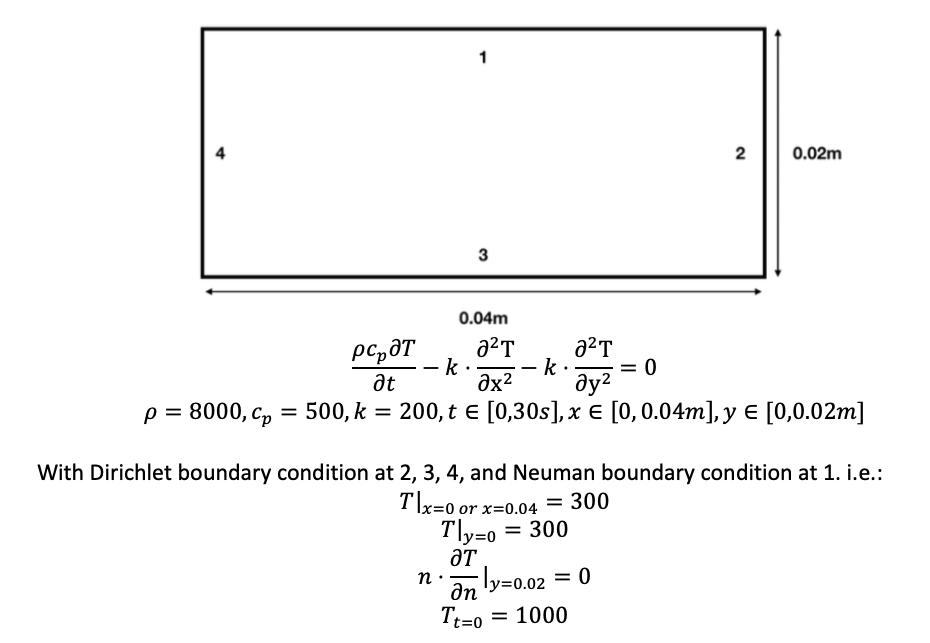
The following is my script:
However, I could not get the converged solution. Basically, the errors on the boundary are not converging. See the following:
Do you have any suggestion how to fix this problem?
Thank you in advance!
The text was updated successfully, but these errors were encountered: