How to hive partitioning into Parquet a VCF format file?
In order to create a database from a VCF file, process partitioning highly speed up futher annotation process. Simply use HOWARD Convert tool with --parquet_partitions option. Format of input and output files can be any avaialbe format (e.g. Parquet, VCF, TSV).
This process is not ressource intensive, but can take a while for huge databases. However, using --explode_infos option require much more memory for huge databases.
INPUT=~/howard/databases/dbsnp/current/hg19/b156/dbsnp.parquet
OUTPUT=~/howard/databases/dbsnp/current/hg19/b156/dbsnp.partition.parquet
PARTITIONS="#CHROM" # "#CHROM", "#CHROM,REF,ALT" (for SNV file only)
howard convert \
--input=$INPUT \
--output=$OUTPUT \
--parquet_partitions="#CHROM" \
--threads=8How to hive partitioning into Parquet a huge VCF format file with all annotations exploded into columns?
Due to memory usage with duckDB, huge VCF file convertion can fail. This tip describe hive partitioning of huge VCF files into Parquet, to prevent memory resource crash.
The following bash script is spliting VCF by "#CHROM" and chunk VCF files with a defined size ("CHUNK_SIZE"). Depending on input VCF file, the number of chunk VCF files will be determined by the content (usually number of annotation within the VCF file). Moreover, Parquet files can also be chunked ("CHUNK_SIZE_PARQUET"). Default options This will ensure a memory usage around 4-5Gb.
Moreover, an additional partitioning can be applied, on one or more specific VCF column or INFO annotation: "None" for no partitioning; "REF,ALT" for VCF only with SNV (e.g. dbSNP); "CLINSIG" for ClinVar database (values such as "Pathogenic, Moslty-Pathogenic"...). Note that partitioning only works for small values (a value lenght will create a too long partition folder), and for not too many values for a column (partition fragments is maximum of 1024). This additional partition can take a while.
In order to create a high-performance database for HOWARD, INFO annotations can be exploded in columns. This option is slower, but generate a Parquet database that will be used by picking needed columns for annotation. Annotations to explode can be chosen with more options.
# Files
VCF=/tmp/my.vcf.gz # Input VCF file
PARQUET=/tmp/my.partition.parquet # Output Partitioned Parquet folder
# Tools
BCFTOOLS=bcftools # BCFTools
BGZIP=bgzip # BGZip
HOWARD=howard # HOWARD
TABIX=tabix # Tabix
# Threads
THREADS=12 # Number of threads
# Param
CHUNK_SIZE=1000000000 # 1000000000 for VCF chunk size around 200Mb
CHUNK_SIZE_PARQUET=10000000 # 10000000 for parquet chunk size around 200Mb
PARQUET_PARTITIONS="None" # "None" for no more partition, "REF,ALT" (SNV VCF) or "REF" or "CLNSIG"...
CONVERT_OPTIONS=" --explode_infos " # Explode INFO annotations into columns
# Create output folder
rm -r $PARQUET
mkdir -p $PARQUET
# Extract header
$BCFTOOLS view -h $VCF --threads $THREADS > $PARQUET/header.vcf
# VCF indexing (if necessary)
if [ ! -e $VCF.tbi ]; then
$TABIX $VCF
fi;
# For each chromosome
for chr in $($TABIX -l $VCF | cut -f1); do
if [ "$chr" != "None" ]; then
echo "# Chromosome '$chr'"
# Create chromosome folder
mkdir -p $PARQUET/#CHROM=$chr;
echo "# Chromosome '$chr' - BCFTools filter and split file..."
$BCFTOOLS filter $VCF -r $chr --threads $THREADS | $BCFTOOLS view -H --threads $THREADS | split -a 10 -C $CHUNK_SIZE - $PARQUET/#CHROM=$chr/ --filter="$BGZIP -l1 --threads=$THREADS > \$FILE.gz";
nb_chunk_files=$(ls $PARQUET/#CHROM=$chr/*.gz | wc -l)
# Convert chunk VCF to Parquet
i_chunk_files=0
for file in $PARQUET/#CHROM=$chr/*.gz; do
# Chunk file to TSV and header
((i_chunk_files++))
chunk=$(basename $file | sed s/.gz$//gi)
echo "# Chromosome '$chr' - Convert VCF to Parquet '$chunk' ($PARQUET_PARTITIONS) [$i_chunk_files/$nb_chunk_files]..."
mv $file $file.tsv.gz;
cp $PARQUET/header.vcf $file.tsv.gz.hdr;
# Convert with partitioning or not
$HOWARD convert --input=$file.tsv.gz --output=$file.parquet $CONVERT_OPTIONS --threads=$THREADS --parquet_partitions="$PARQUET_PARTITIONS" --verbosity=ERROR --chunk_size=$CHUNK_SIZE_PARQUET
# Redirect partitioninf folder if any
if [ "$PARQUET_PARTITIONS" != "" ]; then
rsync -a $file.parquet/ $(dirname $file)/
rm -r $file.parquet*
fi;
# Clean
rm -f $file.tsv*
done;
fi;
done;
# Create header
cp $PARQUET/header.vcf $PARQUET.hdr
# Show partitioned Parquet folder
tree -h $PARQUET
How to aggregate all INFO annotations from multiple Parquet databases into one INFO field?
In order to merge all annotations in INFO column of multiple databases, use a SQL query on the list of Parquet databases, and use STRING_AGG duckDB function to aggretate values.
This will probably work only for small databases.
howard query \
--explode_infos \
--explode_infos_prefix='INFO/' \
--query="SELECT \"#CHROM\", POS, REF, ALT, STRING_AGG(INFO, ';') AS INFO \
FROM parquet_scan('tests/databases/annotations/current/hg19/*.parquet', union_by_name = true) \
GROUP BY \"#CHROM\", POS, REF, ALT" \
--output=/tmp/full_annotation.tsv
head -n2 /tmp/full_annotation.tsv#CHROM POS REF ALT INFO
chr1 69093 G T MCAP13=0.001453;REVEL=0.117;SIFT_score=0.082;SIFT_converted_rankscore=0.333;SIFT_pred=T;SIFT4G_score=0.097;SIFT4G_converted_rankscore=0.392;SIFT4G_pred=T;Polyphen2_HDIV_score=0.0;Polyphen2_HDIV_rankscore=0.029;Polyphen2_HDIV_pred=B;Polyphen2_HVAR_score=0.0;Polyphen2_HVAR_rankscore=0.014;Polyphen2_HVAR_pred=B;LRT_score=0.589;LRT_converted_rankscore=0.056;LRT_pred=N;MutationTaster_score=0.635;MutationTaster_converted_rankscore=0.328;MutationTaster_pred=D;MutationAssessor_score=.;MutationAssessor_rankscore=.;MutationAssessor_pred=.;FATHMM_score=6.74;FATHMM_converted_rankscore=0.005;FATHMM_pred=T;PROVEAN_score=0.27;PROVEAN_converted_rankscore=0.043;PROVEAN_pred=N;VEST4_score=0.12;VEST4_rankscore=0.111;MetaSVM_score=-1.003;MetaSVM_rankscore=0.291;MetaSVM_pred=T;MetaLR_score=0.002;MetaLR_rankscore=0.005;MetaLR_pred=T;MetaRNN_score=0.452;MetaRNN_rankscore=0.666;MetaRNN_pred=T;M_CAP_score=0.001;M_CAP_rankscore=0.022;M_CAP_pred=T;REVEL_score=0.117;REVEL_rankscore=0.332;MutPred_score=0.835;MutPred_rankscore=0.943;MVP_score=0.240;MVP_rankscore=0.236;MPC_score=.;MPC_rankscore=.;PrimateAI_score=.;PrimateAI_rankscore=.;PrimateAI_pred=.;DEOGEN2_score=0.008;DEOGEN2_rankscore=0.072;DEOGEN2_pred=T;BayesDel_addAF_score=-0.359;BayesDel_addAF_rankscore=0.042;BayesDel_addAF_pred=T;BayesDel_noAF_score=-0.754;BayesDel_noAF_rankscore=0.033;BayesDel_noAF_pred=T;ClinPred_score=0.090;ClinPred_rankscore=0.112;ClinPred_pred=T;LIST_S2_score=0.065;LIST_S2_rankscore=0.651;LIST_S2_pred=T;Aloft_pred=.,.,;Aloft_Confidence=.,.,;CADD_raw=0.013;CADD_raw_rankscore=0.049;CADD_phred=2.83;DANN_score=0.602;DANN_rankscore=0.064;fathmm_MKL_coding_score=0.505;fathmm_MKL_coding_rankscore=0.287;fathmm_MKL_coding_pred=D;fathmm_XF_coding_score=0.012;fathmm_XF_coding_rankscore=0.001;fathmm_XF_coding_pred=N;Eigen_raw_coding=-0.958;Eigen_raw_coding_rankscore=0.095;Eigen_PC_raw_coding=-0.968;Eigen_PC_raw_coding_rankscore=0.105;GenoCanyon_score=0;GenoCanyon_rankscore=0.012;integrated_fitCons_score=0.487;integrated_fitCons_rankscore=0.14;integrated_confidence_value=0;LINSIGHT=.;LINSIGHT_rankscore=.;GERP___NR=2.31;GERP___RS=1.36;GERP___RS_rankscore=0.211;phyloP100way_vertebrate=1.139;phyloP100way_vertebrate_rankscore=0.311;phyloP30way_mammalian=0.113;phyloP30way_mammalian_rankscore=0.17;phastCons100way_vertebrate=0.841;phastCons100way_vertebrate_rankscore=0.303;phastCons30way_mammalian=0.552;phastCons30way_mammalian_rankscore=0.281;SiPhy_29way_logOdds=6.575;SiPhy_29way_logOdds_rankscore=0.218;Interpro_domain=.,.;GTEx_V8_gene=.;GTEx_V8_tissue=.;InterVar_automated=Uncertain_significance;PVS1=0;PS1=0;PS2=0;PS3=0;PS4=0;PM1=0;PM2=1;PM3=0;PM4=0;PM5=0;PM6=0;PP1=0;PP2=0;PP3=0;PP4=0;PP5=0;BA1=0;BS1=0;BS2=0;BS3=0;BS4=0;BP1=0;BP2=0;BP3=0;BP4=1;BP5=0;BP6=0;BP7=0
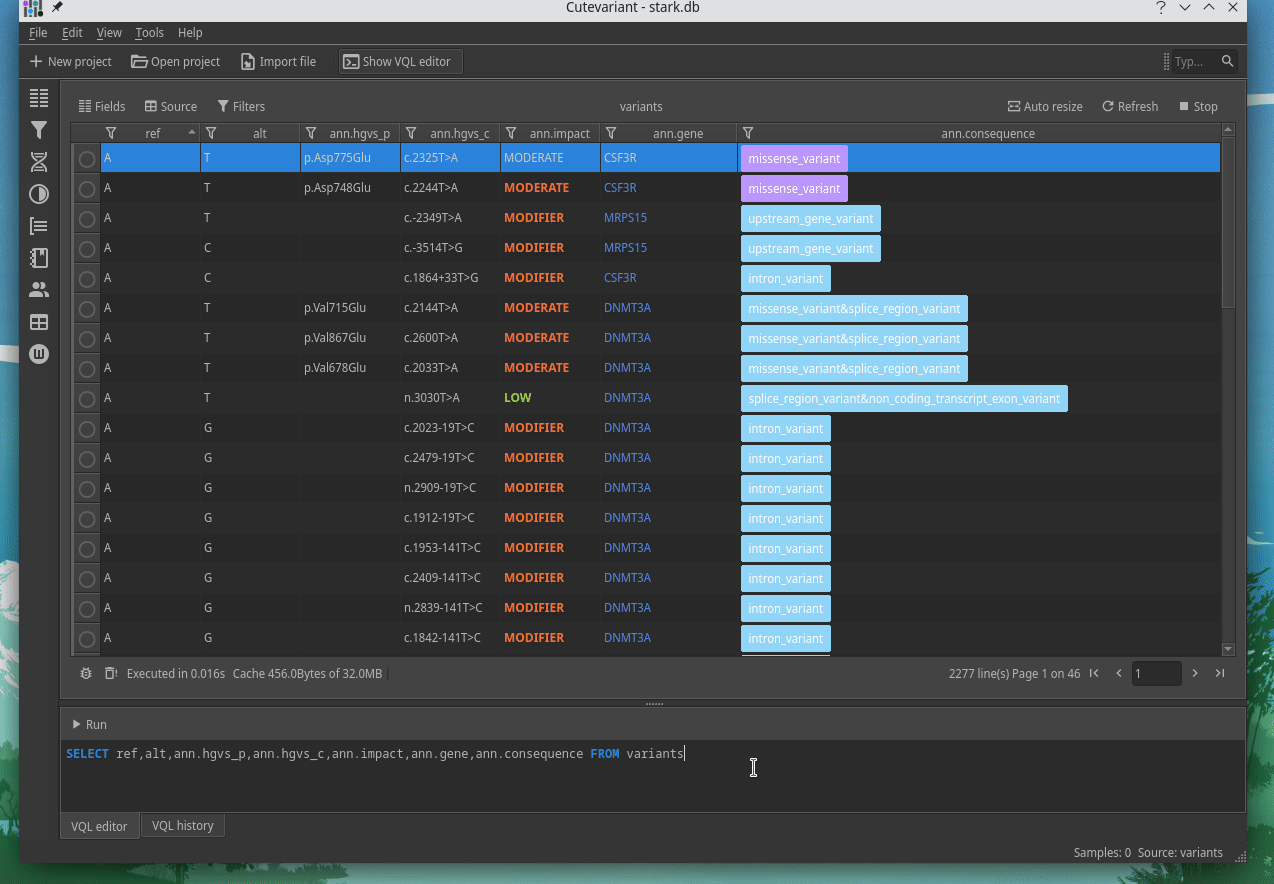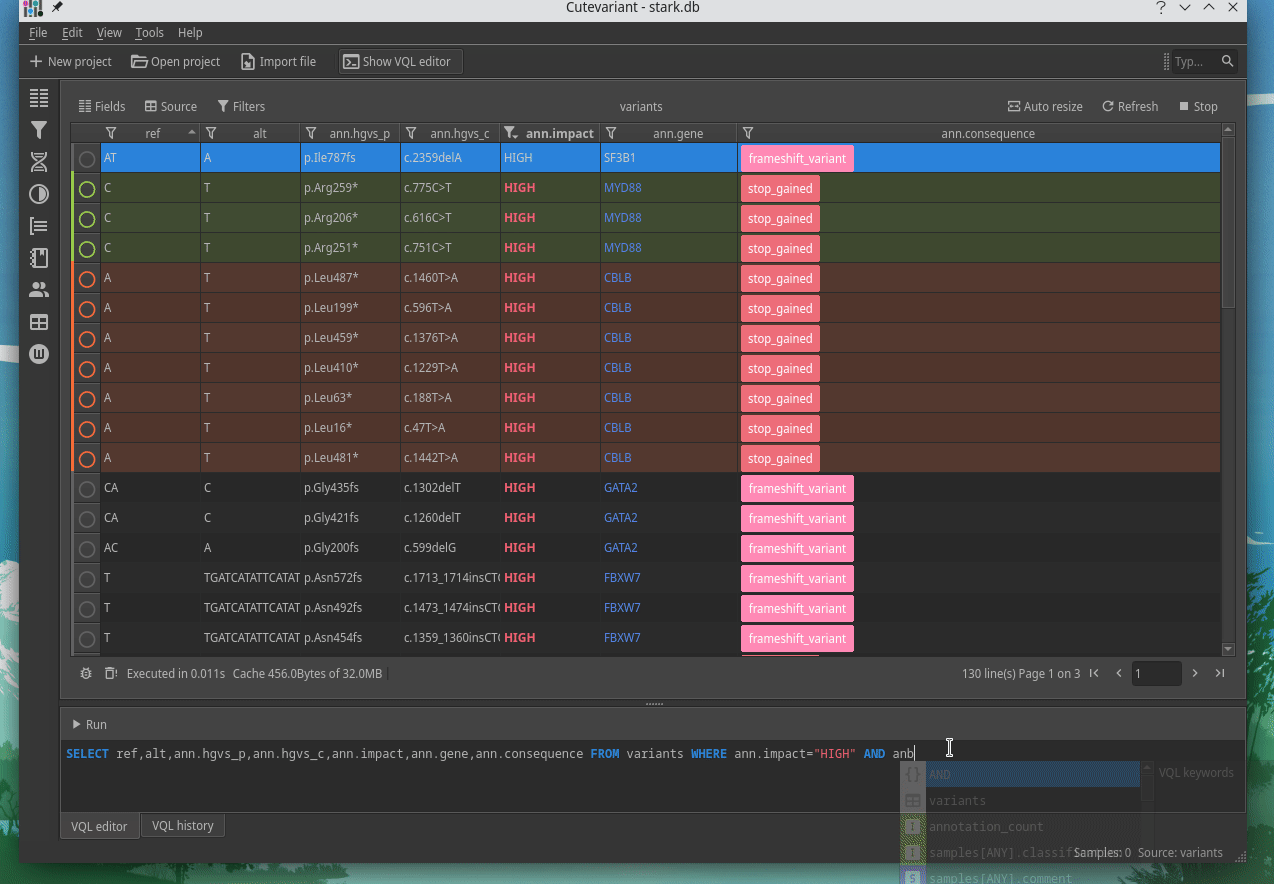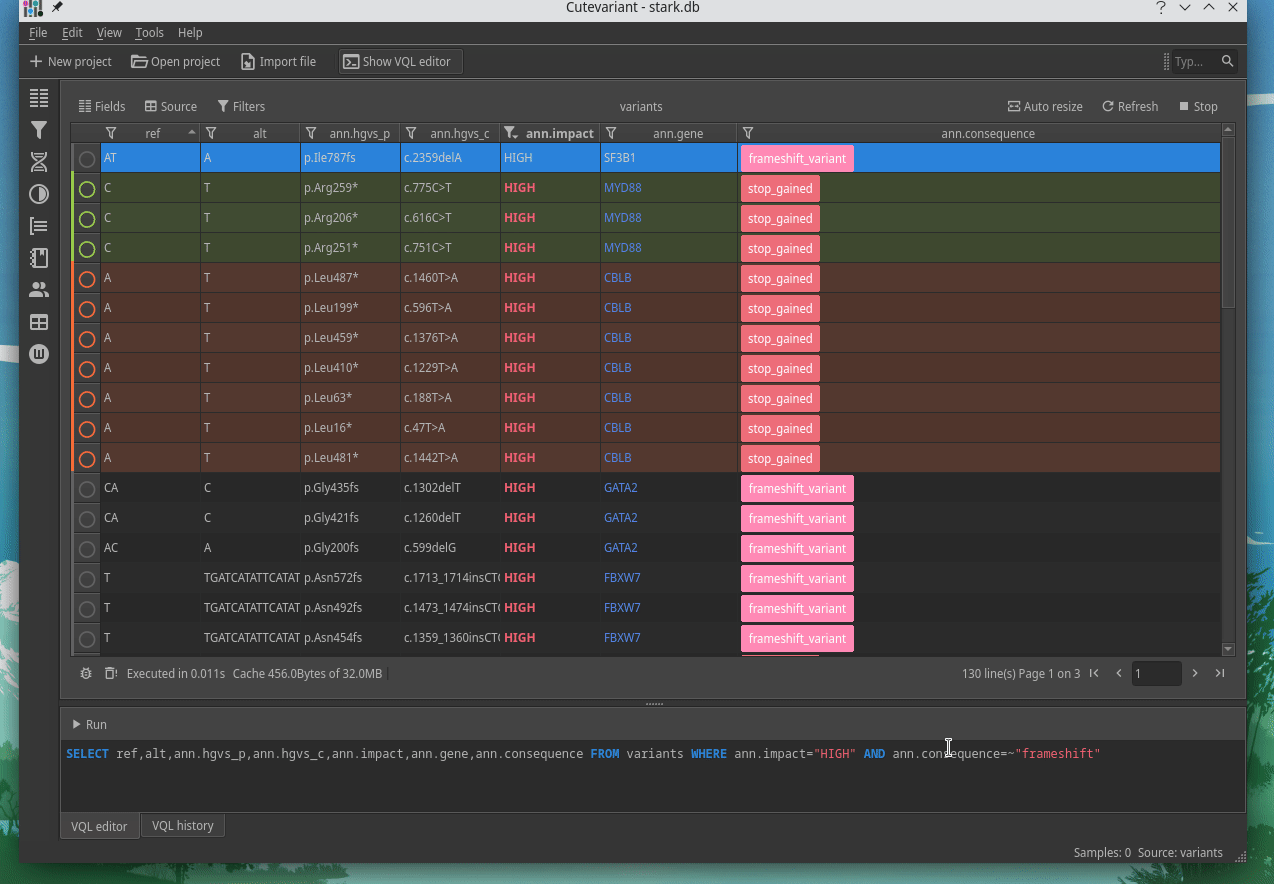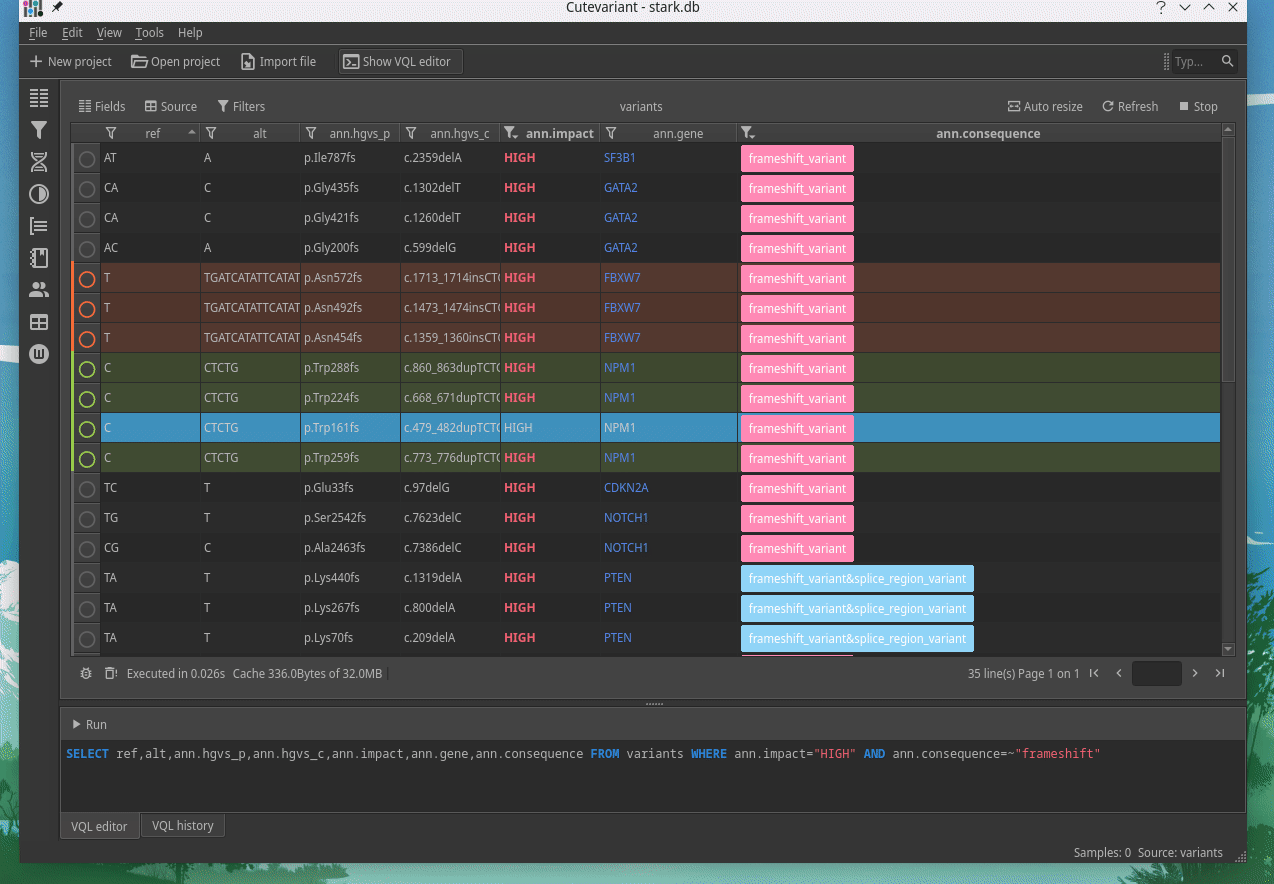
How to explore genetics variations from VCF files?
CuteVariant: A standalone and free application to explore genetics variations from VCF files
Cutevariant is a cross-plateform application dedicated to maniupulate and filter variation from annotated VCF file. When you create a project, data are imported into an sqlite database that cutevariant queries according your needs. Presently, SnpEff and VEP annotations are supported. Once your project is created, you can query variant using different gui controller or directly using the VQL language. This Domain Specific Language is specially designed for cutevariant and try to keep the same syntax than SQL for an easy use.
Documentation available on cutevariant.labsquare.org and GitHub
How to generate dbNSFP databases?
dbNSFP is a database developed for functional prediction and annotation of all potential non-synonymous single-nucleotide variants (nsSNVs) in the human genome.
In order to use dbNSFP with HOWARD, databases need to be downloaded and formatted. The databases tool is able to download and generate VCF, Parquet and Partition Parquet format file.
# Download dbNSFP for ALL annotation, in VCF, Parquet and Partition Parquet, with INFO column
howard databases --assembly=hg19 --download-dbnsfp=~/howard/databases/dbnsfp/4.4a --download-dbnsfp-release='4.4a' --download-dbnsfp-parquet --download-dbnsfp-vcf --download-dbnsfp-add-infoTo generate dbNSFP database in Annovar TXT format, because it can not be provided by Annovar tool, this following script is useful. Itcan be adapted dependgin on the dbNSFP release and needed assembly. For example, for release 4.4a of dbSNFP, and assembly 'hg19', the positional columns are '$8' for chromosome and '$9' for position (see dbNSFP doc).
# dbNSFP GZ to Annovar
# Script index annovar downloaded from https://github.com/WGLab/doc-ANNOVAR/issues/15. Official script does not work with some grep releases (https://gist.githubusercontent.com/suqingdong/447ee784582200309c17b3a844566bac/raw/d227a42ca29caece90bb437a9a10732282d2c35f/index_annovar.pl)
# Usage: perl annovar_idx.pl dbFile binSize
# Example: perl annovar_idx.pl hg19_clinvar_20160302.txt 1000 > hg19_clinvar_20160302.txt.idx
# Init
index_annovar=/path/to/annovar_idx.pl
table_annovar=/path/to/table_annovar.pl
dbSNFP_folder=/path/to/dbnsfp/4.4a/
vcf=/path/to/example.vcf.gz
annovar_file=hg19_dbNSFP44a.txt;
annovar_file_gz=$annovar_file.gz;
# Header
first_file=$(ls $dbSNFP_folder/dbNSFP4.4a_variant.chr*.gz | head -n 1)
nb_column=$(zcat $first_file | head -n1 | awk -F"\t" '{print NF}')
zcat $first_file | head -n1 | awk -v nb_column="$nb_column" -F"\t" '{gsub(/\r/,""); printf "#Chr\tStart\tEnd\tRef\tAlt\t"; for (i = 12; i <= nb_column; i++) {printf "%s\t", $i}; printf "\n"}' > $annovar_file;
echo "Number of column: $nb_column"
# Format each variant file (by chromosome)
for file in $dbSNFP_folder/dbNSFP4.4a_variant.chr*.gz; do
echo $file;
zcat $file | grep "^#" -v | awk -v nb_column="$nb_column" -F"\t" '$8!="."{gsub(/\r/,""); printf $8"\t"$9"\t"$9"\t"$3"\t"$4"\t"; for (i = 12; i <= nb_column; i++) {if($i==""){$i="."}; printf "%s\t", $i}; printf "\n"}' >> $annovar_file;
done;
# Compress
gzip -9 -c $annovar_file > $annovar_file_gz &
# Index
perl $index_annovar $annovar_file 10000 > $annovar_file.idx
# Annovation test
$table_annovar $vcf . -protocol dbNSFP44a_annovar -buildver hg19 -out /tmp/myanno.vcf.gz -remove -operation f -nastring . -vcfinputNote: This script is not parallelized, not optimized