diff --git a/README.md b/README.md
index 594018c..3d3aee8 100644
--- a/README.md
+++ b/README.md
@@ -1,3 +1,4 @@
+
[](https://travis-ci.org/autonlab/DeepSurvivalMachines)
[](https://codecov.io/gh/autonlab/DeepSurvivalMachines)
@@ -6,196 +7,267 @@
[](https://github.com/autonlab/auton-survival)
- +Python package `auton_survival` provides a flexible API for various problems
+in survival analysis, including regression, counterfactual estimation,
+and phenotyping.
+What is Survival Analysis?
+--------------------------
-Package: `auton-survival`
--------------
+**Survival Analysis** involves estimating when an event of interest, \( T \)
+would take places given some features or covariates \( X \). In statistics
+and ML these scenarious are modelled as regression to estimate the conditional
+survival distribution, \( \mathbb{P}(T>t|X) \). As compared to typical
+regression problems, Survival Analysis differs in two major ways:
+* The Event distribution, \( T \) has positive support ie.
+ \( T \in [0, \infty) \).
+* There is presence of censoring ie. a large number of instances of data are
+ lost to follow up.
-Python package `dsm` provides an API to train the Deep Survival Machines
-and associated models for problems in survival analysis. The underlying model
-is implemented in `pytorch`.
+The Auton Survival Package
+---------------------------
-For full documentation of the module, please see https://autonlab.github.io/auton-survival/
+The package `auton_survival` is repository of reusable utilities for projects
+involving censored Time-to-Event Data. `auton_survival` allows rapid
+experimentation including dataset preprocessing, regression, counterfactual
+estimation, clustering and phenotyping and propnsity adjusted evaluation.
-What is Survival Analysis?
-------------------------
-**Survival Analysis** involves estimating when an event of interest, _*T*_
-would take place given some features or covariates _*X*_. In statistics
-and ML, these scenarios are modelled as regression to estimate the conditional
-survival distribution, _*P*_(_T_>t|_X_).
-As compared to typical regression problems, Survival Analysis differs in two major ways:
+Survival Regression
+-------------------
-* The Event distribution, _*T*_ has positive support i.e. _*T*_ ∈ [0, ∞).
-* There is presence of censoring i.e. a large number of instances of data are
- lost to follow up.
+Currently supported Survival Models are:
+### `auton_survival.models.dsm.DeepSurvivalMachines`
+### `auton_survival.models.dcm.DeepCoxMixtures`
+### `auton_survival.models.cph.DeepCoxPH`
-Deep Survival Machines
-----------------------
-
+Python package `auton_survival` provides a flexible API for various problems
+in survival analysis, including regression, counterfactual estimation,
+and phenotyping.
+What is Survival Analysis?
+--------------------------
-Package: `auton-survival`
--------------
+**Survival Analysis** involves estimating when an event of interest, \( T \)
+would take places given some features or covariates \( X \). In statistics
+and ML these scenarious are modelled as regression to estimate the conditional
+survival distribution, \( \mathbb{P}(T>t|X) \). As compared to typical
+regression problems, Survival Analysis differs in two major ways:
+* The Event distribution, \( T \) has positive support ie.
+ \( T \in [0, \infty) \).
+* There is presence of censoring ie. a large number of instances of data are
+ lost to follow up.
-Python package `dsm` provides an API to train the Deep Survival Machines
-and associated models for problems in survival analysis. The underlying model
-is implemented in `pytorch`.
+The Auton Survival Package
+---------------------------
-For full documentation of the module, please see https://autonlab.github.io/auton-survival/
+The package `auton_survival` is repository of reusable utilities for projects
+involving censored Time-to-Event Data. `auton_survival` allows rapid
+experimentation including dataset preprocessing, regression, counterfactual
+estimation, clustering and phenotyping and propnsity adjusted evaluation.
-What is Survival Analysis?
-------------------------
-**Survival Analysis** involves estimating when an event of interest, _*T*_
-would take place given some features or covariates _*X*_. In statistics
-and ML, these scenarios are modelled as regression to estimate the conditional
-survival distribution, _*P*_(_T_>t|_X_).
-As compared to typical regression problems, Survival Analysis differs in two major ways:
+Survival Regression
+-------------------
-* The Event distribution, _*T*_ has positive support i.e. _*T*_ ∈ [0, ∞).
-* There is presence of censoring i.e. a large number of instances of data are
- lost to follow up.
+Currently supported Survival Models are:
+### `auton_survival.models.dsm.DeepSurvivalMachines`
+### `auton_survival.models.dcm.DeepCoxMixtures`
+### `auton_survival.models.cph.DeepCoxPH`
-Deep Survival Machines
-----------------------
-
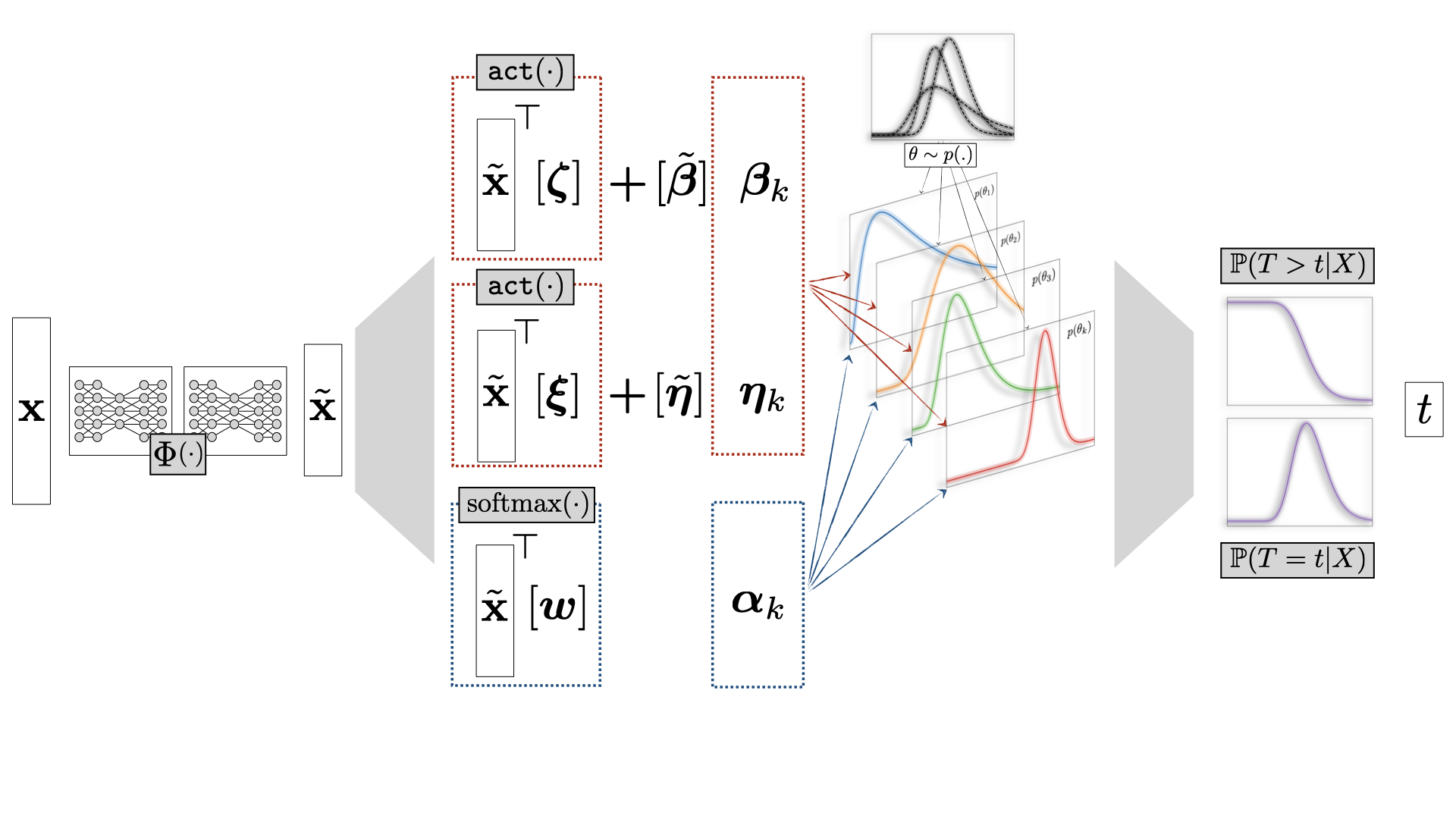 +### `auton_survival.estimators`
+
+This module provids a wrapper to model survival datasets with standard
+survival (time-to-event) analysis methods. The use of the wrapper allows
+a simple standard interface for multiple different survival regression methods.
+
+`auton_survival.estimators` also provides convenient wrappers around other popular
+python survival analysis packages to experiment with the following
+survival regression estimators
+- Random Survival Forests (`pysurvival`):
+- Weibull Accelerated Failure Time (`lifelines`) :
-**Deep Survival Machines (DSM)** is a **fully parametric** approach to model
-Time-to-Event outcomes in the presence of Censoring, first introduced in
-[\[1\]](https://arxiv.org/abs/2003.01176).
-In the context of Healthcare ML and Biostatistics, this is known as 'Survival
-Analysis'. The key idea behind Deep Survival Machines is to model the
-underlying event outcome distribution as a mixure of some fixed \( K \)
-parametric distributions. The parameters of these mixture distributions as
-well as the mixing weights are modelled using Neural Networks.
+### `auton_survival.experiments`
-#### Usage Example
+Modules to perform standard survival analysis experiments. This module
+provides a top-level interface to run `auton_survival` style experiments
+of survival analysis, involving cross-validation style experiments with
+multiple different survival analysis models at different horizons of
+event times.
+
+The module further eases evaluation by automatically computing the
+*censoring adjusted* estimates of the Metrics of interest, like
+**Time Dependent Concordance Index** and **Brier Score** with **IPCW**
+adjustment.
```python
-from dsm import DeepSurvivalMachines
-model = DeepSurvivalMachines()
-model.fit()
-model.predict_risk()
+# auton_survival Style Cross Validation Experiment.
+from auton_survival import datasets
+features, outcomes = datasets.load_topcat()
+
+from auton_survival.experiments import SurvivalCVRegressionExperiment
+
+# instantiate an auton_survival Experiment by
+# specifying the features and outcomes to use.
+experiment = SurvivalCVRegressionExperiment(features, outcomes)
+
+# Fit the `experiment` object with a Cox Model
+experiment.fit(model='cph')
+
+# Evaluate the performance at time=1 year horizon.
+scores = experiment.evaluate(time=1.)
+
+print(scores)
```
-Recurrent Deep Survival Machines
---------------------------------
-
+### `auton_survival.estimators`
+
+This module provids a wrapper to model survival datasets with standard
+survival (time-to-event) analysis methods. The use of the wrapper allows
+a simple standard interface for multiple different survival regression methods.
+
+`auton_survival.estimators` also provides convenient wrappers around other popular
+python survival analysis packages to experiment with the following
+survival regression estimators
+- Random Survival Forests (`pysurvival`):
+- Weibull Accelerated Failure Time (`lifelines`) :
-**Deep Survival Machines (DSM)** is a **fully parametric** approach to model
-Time-to-Event outcomes in the presence of Censoring, first introduced in
-[\[1\]](https://arxiv.org/abs/2003.01176).
-In the context of Healthcare ML and Biostatistics, this is known as 'Survival
-Analysis'. The key idea behind Deep Survival Machines is to model the
-underlying event outcome distribution as a mixure of some fixed \( K \)
-parametric distributions. The parameters of these mixture distributions as
-well as the mixing weights are modelled using Neural Networks.
+### `auton_survival.experiments`
-#### Usage Example
+Modules to perform standard survival analysis experiments. This module
+provides a top-level interface to run `auton_survival` style experiments
+of survival analysis, involving cross-validation style experiments with
+multiple different survival analysis models at different horizons of
+event times.
+
+The module further eases evaluation by automatically computing the
+*censoring adjusted* estimates of the Metrics of interest, like
+**Time Dependent Concordance Index** and **Brier Score** with **IPCW**
+adjustment.
```python
-from dsm import DeepSurvivalMachines
-model = DeepSurvivalMachines()
-model.fit()
-model.predict_risk()
+# auton_survival Style Cross Validation Experiment.
+from auton_survival import datasets
+features, outcomes = datasets.load_topcat()
+
+from auton_survival.experiments import SurvivalCVRegressionExperiment
+
+# instantiate an auton_survival Experiment by
+# specifying the features and outcomes to use.
+experiment = SurvivalCVRegressionExperiment(features, outcomes)
+
+# Fit the `experiment` object with a Cox Model
+experiment.fit(model='cph')
+
+# Evaluate the performance at time=1 year horizon.
+scores = experiment.evaluate(time=1.)
+
+print(scores)
```
-Recurrent Deep Survival Machines
---------------------------------
-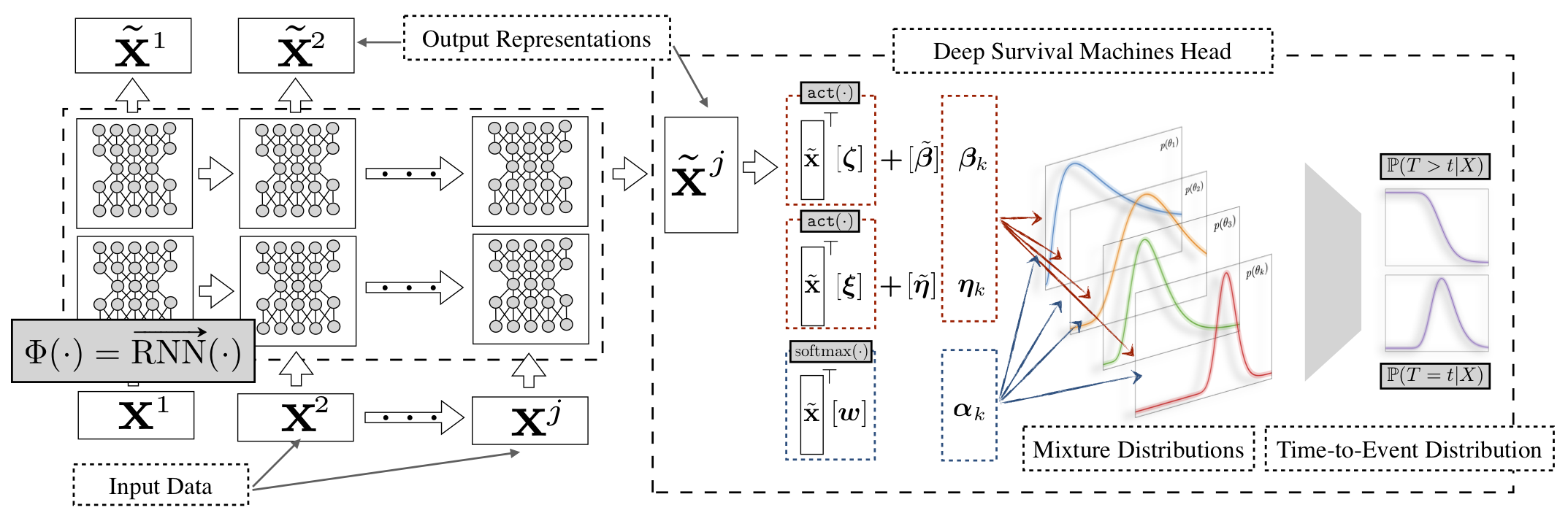 +Phenotyping and Knowledge Discovery
+-----------------------------------
-**Recurrent Deep Survival Machines (RDSM)** builds on the original **DSM**
-model and allows for learning of representations of the input covariates using
-**Recurrent Neural Networks** like **LSTMs, GRUs**. Deep Recurrent Survival
-Machines is a natural fit to model problems where there are time dependendent
-covariates.
+### `auton_survival.phenotyping`
+`auton_survival.phenotyping` allows extraction of latent clusters or subgroups
+of patients that demonstrate similar outcomes. In the context of this package,
+we refer to this task as **phenotyping**. `auton_survival.phenotyping` allows:
-Deep Convolutional Survival Machines
-------------------------------------
+- **Unsupervised Phenotyping**: Involves first performing dimensionality
+reduction on the inpute covariates \( x \) followed by the use of a clustering
+algorithm on this representation.
-Predictive maintenance and medical imaging sometimes requires to work with
-image streams. Deep Convolutional Survival Machines extends **DSM** and
-**DRSM** to learn representations of the input image data using
-convolutional layers. If working with streaming data, the learnt
-representations are then passed through an LSTM to model temporal dependencies
-before determining the underlying survival distributions.
+- **Factual Phenotyping**: Involves the use of structured latent variable
+models, `auton_survival.models.dcm.DeepCoxMixtures` or
+`auton_survival.models.dsm.DeepSurvivalMachines` to recover phenogroups that
+demonstrate differential observed survival rates.
-> :warning: **Not Implemented Yet!**
+- **Counterfactual Phenotyping**: Involves learning phenotypes that demonstrate
+heterogenous treatment effects. That is, the learnt phenogroups have differential
+response to a specific intervention. Relies on the specially designed
+`auton_survival.models.cmhe.DeepCoxMixturesHeterogenousEffects` latent variable model.
-Deep Cox Mixtures
-------------------
-
+Phenotyping and Knowledge Discovery
+-----------------------------------
-**Recurrent Deep Survival Machines (RDSM)** builds on the original **DSM**
-model and allows for learning of representations of the input covariates using
-**Recurrent Neural Networks** like **LSTMs, GRUs**. Deep Recurrent Survival
-Machines is a natural fit to model problems where there are time dependendent
-covariates.
+### `auton_survival.phenotyping`
+`auton_survival.phenotyping` allows extraction of latent clusters or subgroups
+of patients that demonstrate similar outcomes. In the context of this package,
+we refer to this task as **phenotyping**. `auton_survival.phenotyping` allows:
-Deep Convolutional Survival Machines
-------------------------------------
+- **Unsupervised Phenotyping**: Involves first performing dimensionality
+reduction on the inpute covariates \( x \) followed by the use of a clustering
+algorithm on this representation.
-Predictive maintenance and medical imaging sometimes requires to work with
-image streams. Deep Convolutional Survival Machines extends **DSM** and
-**DRSM** to learn representations of the input image data using
-convolutional layers. If working with streaming data, the learnt
-representations are then passed through an LSTM to model temporal dependencies
-before determining the underlying survival distributions.
+- **Factual Phenotyping**: Involves the use of structured latent variable
+models, `auton_survival.models.dcm.DeepCoxMixtures` or
+`auton_survival.models.dsm.DeepSurvivalMachines` to recover phenogroups that
+demonstrate differential observed survival rates.
-> :warning: **Not Implemented Yet!**
+- **Counterfactual Phenotyping**: Involves learning phenotypes that demonstrate
+heterogenous treatment effects. That is, the learnt phenogroups have differential
+response to a specific intervention. Relies on the specially designed
+`auton_survival.models.cmhe.DeepCoxMixturesHeterogenousEffects` latent variable model.
-Deep Cox Mixtures
-------------------
- +Dataset Loading and Preprocessing
+---------------------------------
-The Cox Mixture involves the assumption that the survival function
-of the individual to be a mixture of K Cox Models. Conditioned on each
-subgroup Z=k; the PH assumptions are assumed to hold and the baseline
-hazard rates is determined non-parametrically using an spline-interpolated
-Breslow's estimator.
-For full details on Deep Cox Mixture, refer to the paper:
+Helper functions to load and prerocsss various time-to-event data like the
+popular `SUPPORT`, `FRAMINGHAM` and `PBC` dataset for survival analysis.
-Deep Cox Mixtures
-for Survival Regression. Machine Learning in Health Conference (2021)
+### `auton_survival.datasets`
-Installation
-------------
+```python
+# Load the SUPPORT Dataset
+from auton_survival import dataset
+features, outcomes = datasets.load_dataset('SUPPORT')
+```
+
+### `auton_survival.preprocessing`
+This module provides a flexible API to perform imputation and data
+normalization for downstream machine learning models. The module has
+3 distinct classes, `Scaler`, `Imputer` and `Preprocessor`. The `Preprocessor`
+class is a composite transform that does both Imputing ***and*** Scaling with
+a single function call.
+
+```python
+# Preprocessing loaded Datasets
+from auton_survival import datasets
+features, outcomes = datasets.load_topcat()
+
+from auton_survival.preprocessing import Preprocessing
+features = Preprocessor().fit_transform(features,
+ cat_feats=['GENDER', 'ETHNICITY', 'SMOKE'],
+ num_feats=['height', 'weight'])
+
+# The `cat_feats` and `num_feats` lists would contain all the categorical and
+# numerical features in the dataset.
-```console
-foo@bar:~$ git clone https://github.com/autonlab/auton-survival.git
-foo@bar:~$ cd auton-survival
-foo@bar:~$ pip install -r requirements.txt
```
-Examples
----------
+Evaluation and Reporting
+-------------------------
-1. [Deep Survival Machines on the SUPPORT Dataset](https://nbviewer.jupyter.org/github/autonlab/DeepSurvivalMachines/blob/master/examples/DSM%20on%20SUPPORT%20Dataset.ipynb)
-2. [Recurrent Deep Survival Machines on the PBC Dataset](https://nbviewer.org/github/autonlab/DeepSurvivalMachines/blob/master/examples/RDSM%20on%20PBC%20Dataset.ipynb)
+### `auton_survival.metrics`
+Helper functions to generate standard reports for common Survival Analysis tasks.
-References
-----------
+Citing and References
+----------------------
-Please cite the following papers if you are using the `dsm` package.
+Please cite the following papers if you are using the `auton_survival` package.
[1] [Deep Survival Machines:
Fully Parametric Survival Regression and
-Representation Learning for Censored Data with Competing Risks.
-IEEE Journal of Biomedical \& Health Informatics (2021)](https://arxiv.org/abs/2003.01176)
+Representation Learning for Censored Data with Competing Risks."
+IEEE Journal of Biomedical and Health Informatics (2021)](https://arxiv.org/abs/2003.01176)
```
- @article{nagpal2021deep,
- title={Deep Survival Machines: Fully Parametric Survival Regression and\
- Representation Learning for Censored Data with Competing Risks},
+ @article{nagpal2021dsm,
+ title={Deep survival machines: Fully parametric survival regression and representation learning for censored data with competing risks},
author={Nagpal, Chirag and Li, Xinyu and Dubrawski, Artur},
journal={IEEE Journal of Biomedical and Health Informatics},
- year={2021}
+ volume={25},
+ number={8},
+ pages={3163--3175},
+ year={2021},
+ publisher={IEEE}
}
```
-
-[2] [Deep Parametric Time-to-Event Regression with Time-Varying Covariates.
-AAAI Spring Symposium (2021)](http://proceedings.mlr.press/v146/nagpal21a.html)
+[2] [Deep Parametric Time-to-Event Regression with Time-Varying Covariates. AAAI
+Spring Symposium (2021)](http://proceedings.mlr.press/v146/nagpal21a.html)
```
-@InProceedings{pmlr-v146-nagpal21a,
- title = {Deep Parametric Time-to-Event Regression with Time-Varying Covariates},
- author = {Nagpal, Chirag and Jeanselme, Vincent and Dubrawski, Artur},
- booktitle = {Proceedings of AAAI Spring Symposium on Survival Prediction - Algorithms, Challenges, and Applications 2021},
- series = {Proceedings of Machine Learning Research},
- publisher = {PMLR},
+ @InProceedings{pmlr-v146-nagpal21a,
+ title={Deep Parametric Time-to-Event Regression with Time-Varying Covariates},
+ author={Nagpal, Chirag and Jeanselme, Vincent and Dubrawski, Artur},
+ booktitle={Proceedings of AAAI Spring Symposium on Survival Prediction - Algorithms, Challenges, and Applications 2021},
+ series={Proceedings of Machine Learning Research},
+ publisher={PMLR},
}
```
-[3] [Deep Cox Mixtures for Survival Regression. Machine Learning for Healthcare (2021)](https://proceedings.mlr.press/v149/nagpal21a)
+[3] [Deep Cox Mixtures for Survival Regression. Conference on Machine Learning for
+Healthcare (2021)](https://arxiv.org/abs/2101.06536)
```
-@InProceedings{nagpal2021dcm,
- title={Deep Cox Mixtures for Survival Regression},
+ @inproceedings{nagpal2021dcm,
+ title={Deep Cox mixtures for survival regression},
author={Nagpal, Chirag and Yadlowsky, Steve and Rostamzadeh, Negar and Heller, Katherine},
- booktitle={Proceedings of the 6th Machine Learning for Healthcare Conference},
+ booktitle={Machine Learning for Healthcare Conference},
pages={674--708},
year={2021},
- volume={149},
- series={Proceedings of Machine Learning Research},
- publisher={PMLR},
-}
+ organization={PMLR}
+ }
+```
+
+[4] [Counterfactual Phenotyping with Censored Time-to-Events (2022)](https://arxiv.org/abs/2202.11089)
+
+```
+ @article{nagpal2022counterfactual,
+ title={Counterfactual Phenotyping with Censored Time-to-Events},
+ author={Nagpal, Chirag and Goswami, Mononito and Dufendach, Keith and Dubrawski, Artur},
+ journal={arXiv preprint arXiv:2202.11089},
+ year={2022}
+ }
+```
+
+## Installation
+
+```console
+foo@bar:~$ git clone https://github.com/autonlab/auton_survival
+foo@bar:~$ pip install -r requirements.txt
```
Compatibility
-------------
-`dsm` requires `python` 3.5+ and `pytorch` 1.1+.
+`auton_survival` requires `python` 3.5+ and `pytorch` 1.1+.
To evaluate performance using standard metrics
-`dsm` requires `scikit-survival`.
+`auton_survival` requires `scikit-survival`.
Contributing
------------
-`dsm` is [on GitHub]. Bug reports and pull requests are welcome.
+`auton_survival` is [on GitHub]. Bug reports and pull requests are welcome.
-[on GitHub]: https://github.com/chiragnagpal/deepsurvivalmachines
+[on GitHub]: https://github.com/autonlab/auton-survival
License
-------
MIT License
-Copyright (c) 2020 Carnegie Mellon University, [Auton Lab](http://www.autonlab.org)
-
-Permission is hereby granted, free of charge, to any person obtaining a copy of this
-software and associated documentation files (the "Software"), to deal in the Software
-without restriction, including without limitation the rights to use, copy, modify,
-merge, publish, distribute, sublicense, and/or sell copies of the Software, and to permit
-persons to whom the Software is furnished to do so, subject to the following conditions:
-
-The above copyright notice and this permission notice shall be included in all copies
-or substantial portions of the Software.
-
-THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR IMPLIED,
-INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY, FITNESS FOR A PARTICULAR
-PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE
-FOR ANY CLAIM, DAMAGES OR OTHER LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR
-OTHERWISE, ARISING FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER
-DEALINGS IN THE SOFTWARE.
-
-
+Dataset Loading and Preprocessing
+---------------------------------
-The Cox Mixture involves the assumption that the survival function
-of the individual to be a mixture of K Cox Models. Conditioned on each
-subgroup Z=k; the PH assumptions are assumed to hold and the baseline
-hazard rates is determined non-parametrically using an spline-interpolated
-Breslow's estimator.
-For full details on Deep Cox Mixture, refer to the paper:
+Helper functions to load and prerocsss various time-to-event data like the
+popular `SUPPORT`, `FRAMINGHAM` and `PBC` dataset for survival analysis.
-Deep Cox Mixtures
-for Survival Regression. Machine Learning in Health Conference (2021)
+### `auton_survival.datasets`
-Installation
-------------
+```python
+# Load the SUPPORT Dataset
+from auton_survival import dataset
+features, outcomes = datasets.load_dataset('SUPPORT')
+```
+
+### `auton_survival.preprocessing`
+This module provides a flexible API to perform imputation and data
+normalization for downstream machine learning models. The module has
+3 distinct classes, `Scaler`, `Imputer` and `Preprocessor`. The `Preprocessor`
+class is a composite transform that does both Imputing ***and*** Scaling with
+a single function call.
+
+```python
+# Preprocessing loaded Datasets
+from auton_survival import datasets
+features, outcomes = datasets.load_topcat()
+
+from auton_survival.preprocessing import Preprocessing
+features = Preprocessor().fit_transform(features,
+ cat_feats=['GENDER', 'ETHNICITY', 'SMOKE'],
+ num_feats=['height', 'weight'])
+
+# The `cat_feats` and `num_feats` lists would contain all the categorical and
+# numerical features in the dataset.
-```console
-foo@bar:~$ git clone https://github.com/autonlab/auton-survival.git
-foo@bar:~$ cd auton-survival
-foo@bar:~$ pip install -r requirements.txt
```
-Examples
----------
+Evaluation and Reporting
+-------------------------
-1. [Deep Survival Machines on the SUPPORT Dataset](https://nbviewer.jupyter.org/github/autonlab/DeepSurvivalMachines/blob/master/examples/DSM%20on%20SUPPORT%20Dataset.ipynb)
-2. [Recurrent Deep Survival Machines on the PBC Dataset](https://nbviewer.org/github/autonlab/DeepSurvivalMachines/blob/master/examples/RDSM%20on%20PBC%20Dataset.ipynb)
+### `auton_survival.metrics`
+Helper functions to generate standard reports for common Survival Analysis tasks.
-References
-----------
+Citing and References
+----------------------
-Please cite the following papers if you are using the `dsm` package.
+Please cite the following papers if you are using the `auton_survival` package.
[1] [Deep Survival Machines:
Fully Parametric Survival Regression and
-Representation Learning for Censored Data with Competing Risks.
-IEEE Journal of Biomedical \& Health Informatics (2021)](https://arxiv.org/abs/2003.01176)
+Representation Learning for Censored Data with Competing Risks."
+IEEE Journal of Biomedical and Health Informatics (2021)](https://arxiv.org/abs/2003.01176)
```
- @article{nagpal2021deep,
- title={Deep Survival Machines: Fully Parametric Survival Regression and\
- Representation Learning for Censored Data with Competing Risks},
+ @article{nagpal2021dsm,
+ title={Deep survival machines: Fully parametric survival regression and representation learning for censored data with competing risks},
author={Nagpal, Chirag and Li, Xinyu and Dubrawski, Artur},
journal={IEEE Journal of Biomedical and Health Informatics},
- year={2021}
+ volume={25},
+ number={8},
+ pages={3163--3175},
+ year={2021},
+ publisher={IEEE}
}
```
-
-[2] [Deep Parametric Time-to-Event Regression with Time-Varying Covariates.
-AAAI Spring Symposium (2021)](http://proceedings.mlr.press/v146/nagpal21a.html)
+[2] [Deep Parametric Time-to-Event Regression with Time-Varying Covariates. AAAI
+Spring Symposium (2021)](http://proceedings.mlr.press/v146/nagpal21a.html)
```
-@InProceedings{pmlr-v146-nagpal21a,
- title = {Deep Parametric Time-to-Event Regression with Time-Varying Covariates},
- author = {Nagpal, Chirag and Jeanselme, Vincent and Dubrawski, Artur},
- booktitle = {Proceedings of AAAI Spring Symposium on Survival Prediction - Algorithms, Challenges, and Applications 2021},
- series = {Proceedings of Machine Learning Research},
- publisher = {PMLR},
+ @InProceedings{pmlr-v146-nagpal21a,
+ title={Deep Parametric Time-to-Event Regression with Time-Varying Covariates},
+ author={Nagpal, Chirag and Jeanselme, Vincent and Dubrawski, Artur},
+ booktitle={Proceedings of AAAI Spring Symposium on Survival Prediction - Algorithms, Challenges, and Applications 2021},
+ series={Proceedings of Machine Learning Research},
+ publisher={PMLR},
}
```
-[3] [Deep Cox Mixtures for Survival Regression. Machine Learning for Healthcare (2021)](https://proceedings.mlr.press/v149/nagpal21a)
+[3] [Deep Cox Mixtures for Survival Regression. Conference on Machine Learning for
+Healthcare (2021)](https://arxiv.org/abs/2101.06536)
```
-@InProceedings{nagpal2021dcm,
- title={Deep Cox Mixtures for Survival Regression},
+ @inproceedings{nagpal2021dcm,
+ title={Deep Cox mixtures for survival regression},
author={Nagpal, Chirag and Yadlowsky, Steve and Rostamzadeh, Negar and Heller, Katherine},
- booktitle={Proceedings of the 6th Machine Learning for Healthcare Conference},
+ booktitle={Machine Learning for Healthcare Conference},
pages={674--708},
year={2021},
- volume={149},
- series={Proceedings of Machine Learning Research},
- publisher={PMLR},
-}
+ organization={PMLR}
+ }
+```
+
+[4] [Counterfactual Phenotyping with Censored Time-to-Events (2022)](https://arxiv.org/abs/2202.11089)
+
+```
+ @article{nagpal2022counterfactual,
+ title={Counterfactual Phenotyping with Censored Time-to-Events},
+ author={Nagpal, Chirag and Goswami, Mononito and Dufendach, Keith and Dubrawski, Artur},
+ journal={arXiv preprint arXiv:2202.11089},
+ year={2022}
+ }
+```
+
+## Installation
+
+```console
+foo@bar:~$ git clone https://github.com/autonlab/auton_survival
+foo@bar:~$ pip install -r requirements.txt
```
Compatibility
-------------
-`dsm` requires `python` 3.5+ and `pytorch` 1.1+.
+`auton_survival` requires `python` 3.5+ and `pytorch` 1.1+.
To evaluate performance using standard metrics
-`dsm` requires `scikit-survival`.
+`auton_survival` requires `scikit-survival`.
Contributing
------------
-`dsm` is [on GitHub]. Bug reports and pull requests are welcome.
+`auton_survival` is [on GitHub]. Bug reports and pull requests are welcome.
-[on GitHub]: https://github.com/chiragnagpal/deepsurvivalmachines
+[on GitHub]: https://github.com/autonlab/auton-survival
License
-------
MIT License
-Copyright (c) 2020 Carnegie Mellon University, [Auton Lab](http://www.autonlab.org)
-
-Permission is hereby granted, free of charge, to any person obtaining a copy of this
-software and associated documentation files (the "Software"), to deal in the Software
-without restriction, including without limitation the rights to use, copy, modify,
-merge, publish, distribute, sublicense, and/or sell copies of the Software, and to permit
-persons to whom the Software is furnished to do so, subject to the following conditions:
-
-The above copyright notice and this permission notice shall be included in all copies
-or substantial portions of the Software.
-
-THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR IMPLIED,
-INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY, FITNESS FOR A PARTICULAR
-PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE
-FOR ANY CLAIM, DAMAGES OR OTHER LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR
-OTHERWISE, ARISING FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER
-DEALINGS IN THE SOFTWARE.
-
- -
- +Copyright (c) 2022 Carnegie Mellon University, [Auton Lab](http://autonlab.org)
+
+Permission is hereby granted, free of charge, to any person obtaining a copy
+of this software and associated documentation files (the "Software"), to deal
+in the Software without restriction, including without limitation the rights
+to use, copy, modify, merge, publish, distribute, sublicense, and/or sell
+copies of the Software, and to permit persons to whom the Software is
+furnished to do so, subject to the following conditions:
+
+The above copyright notice and this permission notice shall be included in all
+copies or substantial portions of the Software.
+
+THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
+IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
+FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE
+AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
+LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM,
+OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE
+SOFTWARE.
+
+
+Copyright (c) 2022 Carnegie Mellon University, [Auton Lab](http://autonlab.org)
+
+Permission is hereby granted, free of charge, to any person obtaining a copy
+of this software and associated documentation files (the "Software"), to deal
+in the Software without restriction, including without limitation the rights
+to use, copy, modify, merge, publish, distribute, sublicense, and/or sell
+copies of the Software, and to permit persons to whom the Software is
+furnished to do so, subject to the following conditions:
+
+The above copyright notice and this permission notice shall be included in all
+copies or substantial portions of the Software.
+
+THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
+IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
+FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE
+AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
+LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM,
+OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE
+SOFTWARE.
+
+ +
+![]() +
+
+
+
+
 -
- +Copyright (c) 2022 Carnegie Mellon University, [Auton Lab](http://autonlab.org)
+
+Permission is hereby granted, free of charge, to any person obtaining a copy
+of this software and associated documentation files (the "Software"), to deal
+in the Software without restriction, including without limitation the rights
+to use, copy, modify, merge, publish, distribute, sublicense, and/or sell
+copies of the Software, and to permit persons to whom the Software is
+furnished to do so, subject to the following conditions:
+
+The above copyright notice and this permission notice shall be included in all
+copies or substantial portions of the Software.
+
+THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
+IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
+FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE
+AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
+LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM,
+OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE
+SOFTWARE.
+
+
+Copyright (c) 2022 Carnegie Mellon University, [Auton Lab](http://autonlab.org)
+
+Permission is hereby granted, free of charge, to any person obtaining a copy
+of this software and associated documentation files (the "Software"), to deal
+in the Software without restriction, including without limitation the rights
+to use, copy, modify, merge, publish, distribute, sublicense, and/or sell
+copies of the Software, and to permit persons to whom the Software is
+furnished to do so, subject to the following conditions:
+
+The above copyright notice and this permission notice shall be included in all
+copies or substantial portions of the Software.
+
+THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
+IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
+FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE
+AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
+LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM,
+OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE
+SOFTWARE.
+
+ +
+